0610
Tuning and validation of an image registration procedure for patient-specific SAR simulation
Eros Montin1,2, Giuseppe Carluccio1, Christopher M Collins1, and Riccardo Lattanzi1,2,3
1Center for Advanced Imaging Innovation and Research (CAI2R) Department of Radiology, Radiology Department, New York University Grossman School of Medicine, New York, New York, USA, New York, NY, United States, 2Bernard and Irene Schwartz Center for Biomedical Imaging, Department of Radiology, New York University Grossman School of Medicine, New York, New York, USA, New York, NY, United States, 3Vilcek Institute of Graduate Biomedical Sciences, New York University Grossman School of Medicine, New York, New York, USA, New York, NY, United States
1Center for Advanced Imaging Innovation and Research (CAI2R) Department of Radiology, Radiology Department, New York University Grossman School of Medicine, New York, New York, USA, New York, NY, United States, 2Bernard and Irene Schwartz Center for Biomedical Imaging, Department of Radiology, New York University Grossman School of Medicine, New York, New York, USA, New York, NY, United States, 3Vilcek Institute of Graduate Biomedical Sciences, New York University Grossman School of Medicine, New York, New York, USA, New York, NY, United States
Synopsis
Keywords: Safety, Whole Body
In this article, we build an automatic pipeline for patient-specific assessment of SAR. Given a target MR image and an atlas comprising a reference image and a model, an MR-shaped model can be obtained by registering the AIM with the target MR and applying the resulting displacement fields to the corresponding ABM. The results of the analysis on simulated data showed that the proposed automatic pipeline was reliable and accurate both in terms of SAR distribution and the similarity of the patient-specific models.Introduction
The interaction between the radiofrequency (RF) electric fields and conductive tissues generates heat which can be a source of discomfort or even tissue damage for the patient. Therefore, whole-body and local Specific energy Absorption Rate (SAR) should be monitored throughout the exam. Local SAR and temperature are mainly estimated through numerical simulations [1,2]. The body model (BM) used in the simulation must be as geometrically similar as possible to the actual patient in order to get a reliable patient-specific SAR estimation, different BMs shapes provide different SAR distributions.In this work, we describe how realistic, patient-specific, BMs can be derived from anatomical MR images using atlas-based segmentation [3]. Given a target MR image and an atlas database comprising an image (AIM) and body model (ABM) pairs, an MR-shaped model can be obtained by registering the AIM with the target MR and applying the resulting displacement fields to the corresponding ABM. This BM can be shaped over the target MR and used for patient-specific SAR evaluation[4].Recent developments in the field of image registration demonstrated how this technique can be used to fuse images from complex datasets, even pediatric patients after oncological therapy (5), where the shape and size of the head may change across follow-ups, because of the physiological process of growth and for of the presence of disease-related deformations (such as hydrocephalus or surgical cavities). Therefore, in this article, we tuned the registration parameters of an atlas-based segmentation algorithm with the ultimate goal to create an automatic pipeline for patient-specific assessment of SAR.Material and Methods
Three BM (Male, Duke, and Normann) [6,7,8] were retrieved and for each model, we simulated the images using our block equation solver CAMRIE [9,10] (Figure 1). Furthermore, the images of Norman were simulated with a GRE sequence having TR=1s, and for different TEs, resulting in 5 simulated images (Figure 3).Image registration was performed using a mixed rigid and nonrigid pyramidal approach [11] that, by increasing the transformation degrees of freedom (DoF) from 3 (translation) to 98304 (Bspline), matches the source to the target image (figure 2). The quality of the B-spline transformation model depends on the number of Control Points (CPs) used in the image. A higher number of CP allows more complex patterns to be estimated but decreases the number of neighbor voxels to be used to compute the metrics. Due to the extreme capability to give complex and plausible deformation fields by avoiding regularizations on the deformation field, Scalar Mplus [5] was selected as the similarity metric [12,13].
Scalar Mplus integrates Mutual information (MI) [14] with the scalar product of the gradient of the target and source image: in this way the optimization is regularized indirectly with a spatial descriptor of the matching, the scalar product of the gradient (NGF) that has its maximum when the two vectors point in the same direction such as at a border between two tissues [rNGF]. The relative weights of MI and NGF are set by the regularization term Tau, higher Taus weights the NGF side of the metric more than MI and vice versa [15].
In order to find the best set of CP and Tau, we registered the 2 AIM of Duke (Target) and Male with the 3 AIM of Norman (Source) varying the number of CP and Tau. The quality of the registration was scored by calculating the average Dice coefficient over the muscle region of the target ABM against the deformed one. (Figure 4).
Results and Discussions
From Figure 4 it is possible to see that CP 32 yielded the best results (P<0.05) with an average Dice of 0.65 ± 0.02, while the best Tau was 100 0.65, ± 0.05. Since the best-performing configurations are the two associated with the right extreme of the parameter chosen in the study, we expect a better performance might be reached using even larger Tau values and more CP.Figure 5 shows an example of the SAR simulated with the target ABM and with the registered version of Norman. In case of a perfect registration, the distribution of SAR in the two models should match. The boxplot reports the distribution of SAR in one of the most important tissues of the brain, the gray matter. It is possible to see that 5 of the 6 models were statistically identical to the target SAR distribution in the region.Conclusions
In the study, we demonstrated that it is possible to shape a body model over an MRI with the final aim of patient-specific SAR estimation. Five among the six ABM simulated resulted in similar SAR distribution compared to their target ones, suggesting that the registration was able to match the source models to the target ones.By the tuning phase, it was possible to see that the best configuration on the dataset is obtained when the CP distance is smaller than 4 mm (128 voxel / 32 #CP) and with a value of Tau higher than 100.Thus, In the future, we plan to add more data to our tuning registration by considering platform changes, varying the sequences, and considering larger values of tau and more dense CP distribution of the B-splines transformation.Acknowledgements
This work was performed under the rubric of the Center for Advanced Imaging Innovation and Research (CAI2R, www.cai2r.net), an NIBIB National Center for Biomedical Imaging and Bioengineering (NIH P41 EB017183).References
- Seo, Y., & Wang, Z. J. (2021). Measurement and evaluation of specific absorption rate and temperature elevation caused by an artificial hip joint during MRI scanning. Scientific Reports, 11(1). https://doi.org/10.1038/s41598-020-80828-7
- Malik, S. J., Hand, J. W., Carmichael, D. W., & Hajnal, J. v. (2022). Evaluation of specific absorption rate and heating in children exposed to a 7T MRI head coil. Magnetic Resonance in Medicine, 88(3), 1434–1449. https://doi.org/10.1002/mrm.29283
- Cabezas, M., Oliver, A., Lladó, X., Freixenet, J., & Bach Cuadra, M. (2011). A review of atlas-based segmentation for magnetic resonance brain images. Computer Methods and Programs in Biomedicine,104(3), e158–e177.https://doi.org/https://doi.org/10.1016/j.cmpb.2011.07.015
- Lee J, Carass A, Jog A, Zhao C, Prince JL. Multi-atlas-based CT synthesis from conventional MRI with patch-based refinement for MRI-based radiotherapy planning. Proc SPIE Int Soc Opt Eng. 2017 Feb;10133:101331I. doi: 10.1117/12.2254571. Epub 2017 Feb 24. PMID: 29142336; PMCID: PMC5682626.
- Montin, E., Belfatto, A., Bologna, M., Meroni, S., Cavatorta, C., Pecori, E., Diletto, B., Massimino, M., Oprandi, M. C., Poggi, G., Arrigoni, F., Peruzzo, D., Pignoli, E., Gandola, L., Cerveri, P., & Mainardi, L. (2020). A multi-metric registration strategy for the alignment of longitudinal brain images in pediatric oncology. Medical and Biological Engineering and Computing.
- Dimbylow PJ. “FDTD calculations of the whole-body averaged SAR in an anatomically realistic voxel model of the human body from 1 MHz to 1 GHz.” Physics in Medicine & Biology. 1997 Mar;42(3):479.
- Collins CM, Smith MB. “Calculations of B1 distribution, SNR, and SAR for a surface coil adjacent to an anatomically‐accurate human body model.” Magnetic Resonance in Medicine: An Official Journal of the International Society for Magnetic Resonance in Medicine. 2001 Apr;45(4):692-9.
- Christ A et al. “The Virtual Family—development of surface-based anatomical models of two adults and two children for dosimetric simulations.” Physics in Medicine & Biology. 2009 Dec;55(2):N23
- Montin, E., Giuseppe, C., Christopher M., C., & Lattanzi, R. (2020). CAMRIE – Cloud-Accessible MRI Emulator. 28th Scientific Meeting of the International Society for Magnetic Resonance in Medicine (ISMRM). Virtual Conference, 08-14 August 2020, 1037
- Cao, Z., Oh, S., Sica, C. T., McGarrity, J. M., Horan, T., Luo, W., & Collins, C. M. (2014). Bloch-based MRI system simulator considering realistic electromagnetic fields for calculation of signal, noise, and specific absorption rate. Magnetic Resonance in Medicine, 72(1), 237–247. https://doi.org/10.1002/mrm.24907
- A. Shackleford, N. Kandasamy, and G. C. Sharp, “{O}n developing{B}-spline registration algorithms for multi-core processors,”Phys MedBiol, vol. 55, pp. 6329–6351, Nov. 2010.https://doi.org/10.1007/s11517-019-02109-4Montin, E., Cutrì, E., Spadola, G., Testori, A., Pennati, G., & Mainardi, L. (2015). Tuning of a deformable image registration procedure for skin component mechanical properties assessment. Proceedings of the Annual International Conference of the IEEE Engineering in Medicine and Biology Society, EMBS. https://doi.org/10.1109/EMBC.2015.7319834
- Montin, E., Cutrì, E., Spadola, G., Testori, A., Pennati, G., & Mainardi, L. (2015). Tuning of a deformable image registration procedure for skin component mechanical properties assessment. Proceedings of the Annual International Conference of the IEEE Engineering in Medicine and Biology Society, EMBS. https://doi.org/10.1109/EMBC.2015.7319834
- Cavatorta, C., Meroni, S., Montin, E., Oprandi, M. C., Pecori, E., Lecchi, M., Diletto, B., Alessandro, O., Peruzzo, D., Biassoni, V., Schiavello, E., Bologna, M., Massimino, M., Poggi, G., Mainardi, L., Arrigoni, F., Spreafico, F., Verderio, P., Pignoli, E., & Gandola, L. (2021). Retrospective study of late radiation-induced damages after focal radiotherapy for childhood brain tumors. PLOS ONE, 16(2), e0247748. https://doi.org/10.1371/journal.pone.0247748
- Viola, P., & Wells III, W. M. (1997). Alignment by Maximization of Mutual Information. In International Journal of Computer Vision (Vol. 24, Issue 2).
- Hodneland, E., Lundervold, A., Rørvik, J., & Munthe-Kaas, A. Z. (2014). Normalized gradient fields for nonlinear motion correction of DCE-MRI time series. Computerized Medical Imaging and Graphics, 38(3), 202–210. https://doi.org/https://doi.org/10.1016/j.compmedimag.2013.12.007
Figures
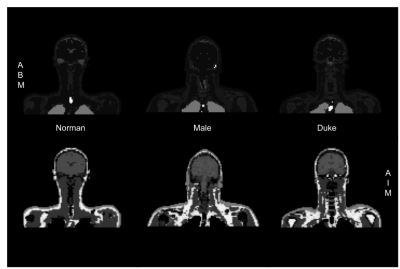
The 3 body models used in the study (ABM) and their synthetic MRI (AIM).
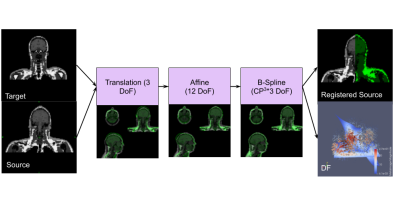
The Registration Workflow, A Target Image (Duke or Mail AIM) is registered to the Norman varying the TE. After that, a 4-layer pyramidal registration concerning 3 deformations is sequentially computed to get the Registered source Image and the Deformation Field associated. The complexity of the deformation increases from 3 Degrees Of Freedom (DoF) of the Translation (Tx, Ty, Tz) up to 98304 in the case of a number of Control Points (CP) of the B-spline equal to 32
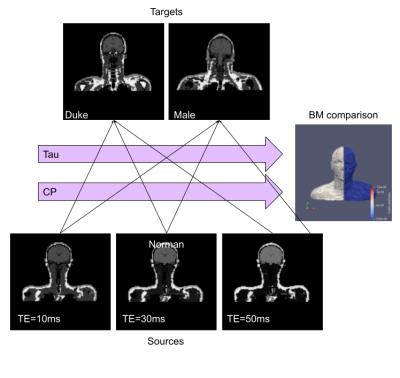
For Every possible parameter of CP and Tau, six registrations are calculated. The DF obtained by the registration is used to warp the Source model and The Dice Coefficient is calculated on the largest Tissue in the model (Muscle)
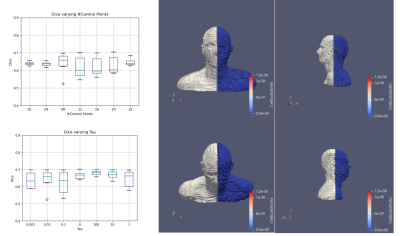
The Results of the analysis. On the left part of the image, the Dice coefficient is shown for varying Tau and the number of Control Points. On the right side, Duke ABM (White) is shown along with Norman Registered ABM (Blue)
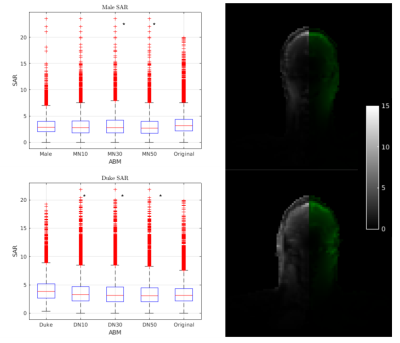
) the box plot of the Grey Matter SAR in the Original Duke (Top) and Male (Bottom) in comparison with the Norman model at different TE after the registration and the original ABM.DN30 and DN50 have an identical distribution of SAR compared to the Original Duke as well as MN10, MN30, and MN50 with Male. Five among the six models after registration have a nearly identical distribution of SAR compared to their Target simulation.
DOI: https://doi.org/10.58530/2023/0610