0577
Ultra-high spatial resolution for ex-vivo structural brain MRI1Core Facility Small Animal Imaging (CF-SANI), University of Ulm, Ulm, Germany, 2German Center for Neurodegenerative Diseases (DZNE), Ulm, Germany
Synopsis
Keywords: Neurodegeneration, Preclinical, ultra-high resolution neuro neuroimaging
This study aims at demonstrating the feasibility of ex vivo MRI for structural imaging of brain. High spatial resolution in the range of 15-20 µm is obtained by an optimized FLASH sequence together with a cryogenically cooled RF coil. A 15µm-resolution was obtained for the brain, revealing cortical grey matter lamination, white matter and vascular architecture. As proof of concept, we showed in an Amyotrophic Lateral Sclerosis ( FUSΔNLS) mouse the pattern of atrophy as proxy of vulnerability using high degree of anatomical granularity. Overall, we demonstrate an ex-vivo MRI strategy with histology-grade resolution, comprehensive and non-destructive brain sampling.Purpose:
Current volumetric imaging of the brain and spinal cord requires either serial sectioning, followed by the painstaking and error-prone re-alignment and reconstruction of image sequences (1), or single-plane illumination microscopy in cleared specimens, which require a time-consuming procedure involving organic solvents or the embedding in hydrogels (2,3). MRI imaging provides a suitable non-destructive platform for the volumetric imaging of intact brain and spinal cord with the advantage of intrinsic contrast (i.e., not necessitating of staining procedures) and speed of acquisition (several hours compared to several days or weeks for histological procedures). Furthermore, the MRI imaging would leave the sample available for additional investigations, such as focused immunohistological studies. However, so far it has been impossible to achieve an ex-vivo MRI resolution comparable to the histological benchmark. Here we demonstrate that using a cryogenically cooled RF coil together with optimized imaging parameters is able to provide high spatial resolution comparable to what is possible with microscopy imaging. We apply this concept at the comparison of brain architecture in WT(Fus+/+) and FusΔNLS mouse (4), a model Amyotrophic Lateral Sclerosis/Frontotemporal Dementia. We also provide proof-of-concept of the application of the ex-vivo imaging to spinal cord, achieved the detection of single groups of motoneurons.Methods:
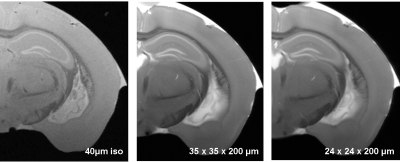
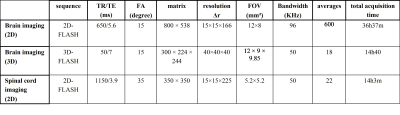
C57/B6 mice were transcardially perfused with PBS (1.5 ml/g) and then fixed with PFA (4% in PBS, 1.5 ml/g). The brain and spinal cord were extracted, post-fixed in PFA (4%) for 18h, washed in PBS and stored in Fluorinert at 4°C. Magnetic resonance imaging (MRI) was performed on a dedicated ultrahigh field 11.7T small animal system (BioSpec 117/16, Bruker Biospin, Ettlingen, Germany) equipped with a 9 cm gradient insert (BGA-S9) operating with ParaVision 6.01. All data were acquired using a cryogenically cooled 1H two-element surface transmit/receive coil (MRI CryoProbe™, Bruker BioSpec, Ettlingen, Germany). All sequence parameters were empirically optimized to provide highest contrast through the brain to delineate maximum substructure and layers. Motion averaging and fat suppression modules were activated in all employed scans. The parameters of each employed sequence were optimized and summarized in Table 1. For the quantification, a dedicated image analysis pipeline was established: the anatomical plate deck from the Allen Brain Atlas was resliced to match the angle of the MRI dataset. Using the nearest plate matching each MRI axial section, the WholeBrain suite was implemented in R-studio and the anatomical landmarks were manually located on each plate. The custom-warped atlas plate was then used to measure volume in each MRI section and provided automated anatomical annotation for 3 levels of anatomical granularity (e.g, “isocortex”, “motor cortex” “primary motor cortex”).Results:
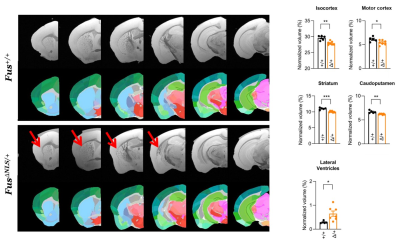
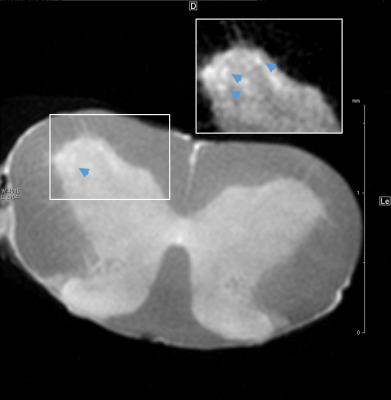
The comparison of WT and FUSΔNLS mice was performed at different level of anatomical granularity. At the largest scale, we detected an atrophy of isocortex, hippocampus, hypothalamus and deep cerebral nuclei, despite no change in overall brain volume. This was justified by a significant increase in ventricular volume, in particular of lateral ventricles. Analysis of cortical areas revealed isolated atrophy of motor cortex, coherent with the features associated with ALS/FTD, but also of Somatosensory cortex and Retrosplenial cortex. Analysis of Basal ganglia revealed an atrophy of Caudoputamen and, more detailed, of the ventral striatum (figure 2). We provide a proof of concept of the imaging of the cervical spinal cord with ex-vivo MRI (figure 3) in FUSΔNLS mice. The resolution of 15µm (figure 3) allows not only the precise separation of white and grey matter boundaries but, in the ventral horn, the idntification of structures compatible with single motoneuron pools (or columns) and possible, for the largest MN with a diameter of >200µm, also of single MN.Discussion:
Our enhanced, ex-vivo MRI imaging paradigm provides an x-y resolution comparable to a histological section imaged in an epifluorescence microscope, with a z thickness also comparable to the thickness used in most immunohistological preparations. The intrinsic contrast was not only sufficient to resolve different cortical layers (layer I, II/III and IV clearly stand out), but also to resolve sub-structures in the corpus callosum, individual vessels and choroid plexuses. Some of these structures are often damaged or warped during sectioning, thus the volumetric imaging of intact brains provides an unprecedented entry point to the study of their intact architecture. When applied to the FUSΔNLS mouse model, the ultra-high spatial resolution allows the characterization of atrophy pattern that reveals involvement beyond what previously demonstrated (4): in fact, the high-resolution ex-vivo MRI is free of the deformation applied during histological preparations and allows the continuous sampling of the brain, instead of relying on discontinuous sections. The proof of concept in the spinal cord, with the definition of what may be single motoneurons column, provide an outlook to the definition of atrophy patterns also in this structure, in particular in ALS.Acknowledgements
DFG grant no. 251293561 to FRReferences
1. Pivetta C, Esposito MS, Sigrist M, Arber S. Motor-circuit communication matrix from spinal cord to brainstem neurons revealed by developmental origin. Cell. 2014;156(3):537-48.
2. Ueda HR, Ertürk A, Chung K, Gradinaru V, Chédotal A, Tomancak P, Keller PJ. Tissue clearing and its applications in neuroscience. Nat Rev Neurosci. 2020;21:61-79.
3. Epp JR, Niibori Y, Liz Hsiang HL, Mercaldo V, Deisseroth K, Josselyn SA, Frankland PW. Optimization of CLARITY for Clearing Whole-Brain and Other Intact Organs. eNeuro. 2015;2:ENEURO.0022-15.2015.
4. Scekic-Zahirovic J, Oussini HE, Mersmann S, Drenner K, Wagner M, Sun Y, Allmeroth K, Dieterlé S, Sinniger J, Dirrig-Grosch S, René F, Dormann D, Haass C, Ludolph AC, Lagier-Tourenne C, Storkebaum E, Dupuis L. Motor neuron intrinsic and extrinsic mechanisms contribute to the pathogenesis of FUS-associated amyotrophic lateral sclerosis. Acta Neuropathol. 2017 Jun;133(6):887-906.
Figures