0569
AMU7T: a 3D qT1 and T2*w high-resolution in vivo template with refined white and gray matter parcellation dedicated to 7T spinal cord MR analyses1Aix Marseille Univ, CNRS, CRMBM, Marseille, France, 2APHM, CHU Timone, Pôle d’Imagerie Médicale, CEMEREM, Marseille, France, 3iLab-Spine, International Associated Laboratory, Marseille-Montreal, France, 4NeuroPoly Lab, Institute of Biomedical Engineering, Polytechnique Montréal, Montréal, QC, Canada
Synopsis
Keywords: Spinal Cord, Software Tools
Ultra-High-Field MRI has opened new perspectives for spinal cord exploration due to improved spatial resolution and contrast. The present work proposes a dedicated 7T multimodal 3D qT1 and T2*w template and a parcellation including eight substructures within gray matter, thirty WM tracts and three inter-hemispheric ROIs, for an accurate atlas-based segmentation in the subject space. This atlas was interpolated in the 3D PAM50 space to benefit from the advanced functions for registration implemented in the SCT. A preliminary segmentation result in healthy subject gives promising perspectives for group studies.INTRODUCTION
The emergence of 7T scanners and the development of quantitative MRI sequences at such field (such as MP2RAGE) have opened new perspectives for the exploration of the spinal cord1,2. However, due to improved spatial resolution and contrasts, conventional tools to analyze the images may fail.Recently, a study proposed by Massire3 highlighted the relevance of quantitative T1 by proposing a first exploratory 7T MRI template (relying on an averaging of ten subjects) revealing anatomical details and facilitating the delineation of WM tracts and GM regions boundaries, in agreement with existing in-vivo4/ex-vivo5 atlases and anatomical drawing illustrated in different books6,7.
The main utility of this atlas lies in the segmentation approach which accurately characterizes (in the subject space) the central canal, the anterior fissure and the posterior septum (that may contaminate T1 quantification if not considered). The atlas also includes five substructures within gray matter horns that could be used to target specifically relevant areas for pathological studies or fMRI. On the other hand, the small number of subjects but also the perfectible template construction method, as well as the lack of spatial coherence related of a 2D registration on data initially acquired in 3D, greatly limited its applicability.
To overcome these limitations, the present work consolidates and completes this atlas, and extends it in the 3D PAM50 space8 using an optimized pipeline for multimodal template construction.
METHODS
This work relies on 2 datasets previously acquired on a 7T MR system (Siemens Healthcare, Germany) with an 8Tx/8Rx neck coil (Rapid Biomedical, Germany) used in CP-mode.The first dataset (AMU26) was composed of transverse 3D-MP2RAGE quantitative T1 maps acquired on 26 healthy subjects (HC) (cf.Table1A and Fig.1.1a)
The second dataset (AMU72), recently proposed for a realistic data augmentation process for automated GM segmentation9) was composed of 2D axial multi-echo GRE T2*-weighted images (T2*w) acquired on 72 subjects (34 HC, 25 Amyotrophic Lateral Sclerosis and 13 Multiple Sclerosis patients). (cf.Table1A and Fig.1.1b).
Preprocessing: All images (N=1208) were automatically segmented with the SCT10 functions sct_deepseg_sc11 and sct_deepseg_gm12,9. Cord/GM masks were manually corrected, and images/masks were Left/Right symmetrized (Fig.1.2) to increase SNR.
Construction of 2D T2*w and qT1 HR templates: Multi-stages SyGN13 processes (Fig.1.2) using SyN transformation were then performed for each level (cf. Table1B-1 for detailed parameters) using either 2D multiple-stacks of qT1 images for the 2D AMU26qT1 template or T2*w image stack for the 2D AMU72T2*w template. For both datasets, the corresponding probabilistic AMU26GM,SC,WM and AMU72GM,SC,WM atlases were generated to constraint the SyGN processes.
To progressively refine the 2D AMU26qT1 template details (Fig.1.3a), 3 different stages were finally performed using the (GM,SC,qT1) modalities (cf. Table1B-2 for detailed parameters). For AMU72T2*w (Fig.1.3b), one single stage was performed using the (GM,SC,T2*w) modalities (cf. Table1B-3).
Generation of the 3D multimodal template AMU7T compatible with PAM50: The 3D propagation of both 2D AMU72T2*w and AMU26qT1 templates into the PAM50 space7,14 was performed step-wise using a set of 2D slice-wise co-registrations (Fig.2.1a):
- i) from AMU26 to AMU72, and (WM30tracts, GM6parcels)4 to AMU72 (cf. details on Table1B-4),
- ii) from AMU72 to PAM50 (cf. Table1B-5).
For each 2D inter-level slice of AMU72 registered to PAM50, 3D interpolations (combining ascending and descending fields of deformations, described previously by Ogier15) were applied (Fig.2.1b) to generate the final 3D multimodal AMU7T template integrating (AMU26qT1,masks, AMU72T2*masks, WM30Tracts, GM16Parcels, , IH3ROI) at (0.175x0.175x5)mm3 resolution.
The GM/WM parcellation (Fig.2.1c) aimed at refining the probabilistic GM6Parcels currently available in SCT (R/L anterior, intermediate and dorsal horns) in a new GM16Parcels parcellation to which is added a subdivision in 3 regions of interest from the inter-hemispheric zone called IH3ROI. These latter, which are in agreement with 3,5, were defined to take advantages of 7T high-resolution and investigate new areas such as motoneuron clusters, while avoiding areas filled with CSF that may contaminated quantification.
RESULTS
A qualitative comparison between PAM50 atlas, ex-vivo atlas and our new AMU7T atlas is highlighted on Fig2.2.a-c.A more quantitative and preliminary result is shown on Fig.3. Individual 3D qT1 quantification obtained after applying the sct_register_to_template function alone (Fig.3.1a, using AMU7T_qT1 template for the registration, and default parameters for other options), or in addition to an optimized SyN registration (Fig.3.1b), demonstrates the feasibility of using this parcellation to accurately segment a 3D qT1 volume in the subject space. Fig.3.2 also shows the impact of registration (SCT or SCT+SyN) on the mean/stdev qT1 measurements extracted from the AMU7T parcels (less bias due to GM/WM and WM/CSF PVE).
CONCLUSION
In line with recent works and progresses in the field of 7T spinal cord MRI, we proposed a new multimodal 3D template (AMU7T) dedicated to 7T qT1 analyses.On the short term, we aim at making it fully compatible with the Spinal Cord Toolbox (SCT10), so as to benefit from the dedicated functions for 3D atlas-based segmentation (template-to-subject registration including straightening, vertebral alignment and slice-wise non-linear registration), with the perspective to improve the reliability and significance of the cluster’s localization extracted by voxel-based analyses in group studies.
Acknowledgements
This work was supported by ARSEP, Fondation Latran and A*midex. The authors sincerely thank R. Dintrich and S. Demortière for SC/GM T2*w manual segmentation and L.Pini, C. Costes and V. Gimenez for study logistic.References
1. Barry, R. L., Vannesjo, S. J., By, S., Gore, J. C. & Smith, S. A. Spinal cord MRI at 7T. Neuroimage 168, 437–451 (2018).
2. Callot, V., Combes, A., Destruel, A. & Smith, S. Ultra-high Field Spinal Cord MRI. in Ultra-high Field Neuro MRI, Elsevier Inc. S&T Books (Amsterdam, NL), in press (2022).
3. Massire, A., Rasoanandrianina, H., Guye, M. & Callot, V. Anterior fissure, central canal, posterior septum and more: New insights into the cervical spinal cord gray and white matter regional organization using T1 mapping at 7T. Neuroimage 205, 116275 (2020).
4. Lévy, S. et al. White matter atlas of the human spinal cord with estimation of partial volume effect. NeuroImage 119, 262–271 (2015).
5. Gros, C. et al. Ex vivo MRI template of the human cervical cord at 80μm isotropic resolution. Proceedings of the 28th Annual Meeting of ISMRM (2020).
6. Hausman, L. Atlases of the Spinal Cord and Brainstem and the Forebrain. (Thomas, 1962).
7. Khan, Y. S. & Lui, F. Neuroanatomy, Spinal Cord. in StatPearls (StatPearls Publishing, 2022).
8. De Leener, B. et al. PAM50: Unbiased multimodal template of the brainstem and spinal cord aligned with the ICBM152 space. NeuroImage 165, 170–179 (2018).
9. Medina, N. J. L., Gros, C., Cohen-Adad, J., Callot, V. & Le Troter, A. 2D Multi-Class Model for Gray and White Matter Segmentation of the Cervical Spinal Cord at 7T. (2021) doi:10.48550/ARXIV.2110.06516.
10. De Leener, B. et al. SCT: Spinal Cord Toolbox, an open-source software for processing spinal cord MRI data. Neuroimage 145, 24–43 (2017).
11. Gros, C. et al. Automatic segmentation of the spinal cord and intramedullary multiple sclerosis lesions with convolutional neural networks. NeuroImage 184, 901–915 (2019).
12. Perone, C. S., Calabrese, E. & Cohen-Adad, J. Spinal cord gray matter segmentation using deep dilated convolutions. Sci Rep 8, 5966 (2018).
13. Avants, B. B. et al. The optimal template effect in hippocampus studies of diseased populations. NeuroImage 49, 2457–2466 (2010).
14. Fonov, V. S. et al. Framework for integrated MRI average of the spinal cord white and gray matter: The MNI-Poly-AMU template. Neuroimage 102, 817–827 (2014).
15. Ogier, A., Sdika, M., Foure, A., Le Troter, A. & Bendahan, D. Individual muscle segmentation in MR images: A 3D propagation through 2D non-linear registration approaches. in 2017 39th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC) 317–320 (IEEE, 2017). doi:10.1109/EMBC.2017.8036826.
16. Taso, M. et al. Construction of an in vivo human spinal cord atlas based on high-resolution MR images at cervical and thoracic levels: preliminary results. Magn Reson Mater Phy 27, 257–267 (2014).
17. Massire, A. et al. High-resolution multi-parametric quantitative magnetic resonance imaging of the human cervical spinal cord at 7T. NeuroImage 143, 58–69 (2016).
18. Taso, M. et al. A reliable spatially normalized template of the human spinal cord--Applications to automated white matter/gray matter segmentation and tensor-based morphometry (TBM) mapping of gray matter alterations occurring with age. Neuroimage 117, 20–28 (2015).
Figures
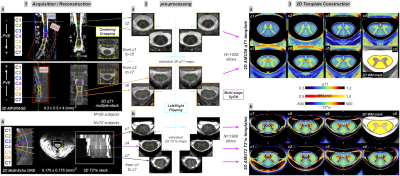
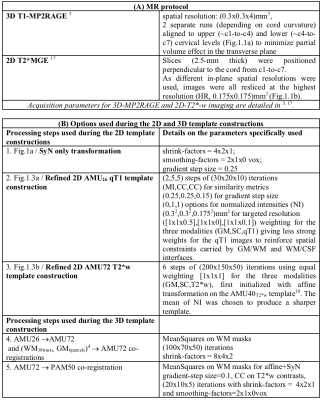
Fig.1 : 2D template construction steps
1. (a) 3D MP2RAGE and (b) 2D T2*w acquisitions. 2. Cord and GM segmentation, followed by L/R flipping (blue color). Multi-stage SyGN13 processes using SyN transformation were then performed (pink color), for each level, using stacks of qT1 images (composed of multiple images per level for each individual subject) for the 2D AMU26qT1 template, and sets of T2*w image stack for the 2D AMU72T2*w template.
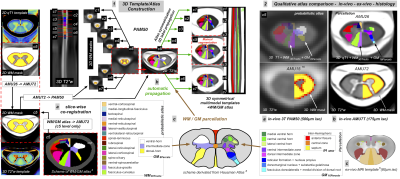
Fig.2 : Post-processing pipeline for the generation of 3D AMU7T atlas
1. (a) slice-wise co-registration steps between AMU spaces and PAM50 (b) automatic 3D label propagation through 2D fields of deformations (c) scheme and legend of WM/GM parcellation 2. Qualitative atlas comparison - in-vivo - ex-vivo - histology (a) in-vivo 3T PAM50 (500μm iso) (b) in-vivo AMU7T (175μm iso) (c) ex-vivo MRI template (80μm iso)
Fig.3 : Qualitative and quantitative results of an automated AMU7T segmentation on one single subject – impact of registration bias
1. on left: sagittal and coronal views of 3D MP2RAGE and labeled mask of cervical levels, on right: axial views at different levels (c2,c4,c6) of AMU7T automated segmentations (a) from SCT default registration (b) from SCT + optimized SyN registration 2. boxplot representation of mean/stdev qT1 values for each hemisphere (a) from SCT default registration (b) from SCT + optimized SyN registration