0557
Deep learning-based Motion-corrected Image Reconstruction for High-resolution Spiral First-pass Myocardial Perfusion Imaging1Biomedical Engineering, University of Virginia, Charlottesville, VA, United States, 2Cardiovascular Medicine, Stanford University, Stanford, CA, United States
Synopsis
Keywords: Heart, Perfusion
Cardiovascular magnetic resonance (CMR) is susceptible to motion-induced artifacts from cardiac and respiratory motion, leading to poor image quality. The inter-frame motion artifacts make quantitative analysis for cardiac function evaluation difficult. Hence motion correction is an important pre-processing step before robust quantification of myocardial perfusion. We developed a deep learning-based framework for rapid and accurate motion correction of CMR perfusion imaging using a 2D U-Net that estimates the deformation field from a moving frame to a fixed frame.
Introduction
About 20.1 million adults aged 20 and older have coronary artery disease (about 7.2%)1. First-pass contrast-enhanced myocardial perfusion imaging provides important diagnostic and prognostic information in coronary artery disease2. High-resolution spiral perfusion imaging techniques, using a motion-compensated L1-SPIRiT reconstruction, are capable of whole-heart high-resolution perfusion imaging3, but the motion-compensated reconstruction is performed off-line and is a time-consuming method, taking ~40 minutes per slice. To address this limitation, we developed a deep learning-based image reconstruction and motion-correction framework, to provide fast and accurate image reconstruction and registration for spiral first-pass myocardial perfusion imaging.Methods
High-resolution spiral perfusion images were reconstructed with a previously proposed rapid image reconstruction network (DESIRE)4 followed with a deep learning-based rapid motion correction. The network structures for image reconstruction and motion correction are demonstrated in Figure 1A and Figure 1B, respectively. A 2D U-Net based network structure was utilized for the estimation of deformation fields. During the training process, image pairs in the perfusion dynamic series are randomly selected, and the network learns the deformation mapping from one frame to the other (moving frame to fixed frame) without a gold-standard image registration. The output of the network is the estimated deformation field between the moving and fixed frames. Motion correction is conducted by applying the deformation field on the moving frame.To test the proposed technique, 210 slices from 35 patients undergoing clinically ordered stress CMR studies with gadolinium (Gd)-based contrast agents on 3T scanners (SIEMENS Prisma/Skyra, Siemens Healthineers, Erlangen, Germany) were used. Datasets were resized to 192 x 192 x 40 (frames). Spiral whole-heart perfusion images had a 1.25 mm in-plane resolution and 10 mm slice thickness3. Images were firstly reconstructed using the previously trained DESIRE framework where complex-valued convolution was enforced4.
For the motion correction network, data from 30 subjects were used for training and data from another 5 subjects were used for validation. Each subject case had four or six slices. Training and evaluation were conducted on a single GPU (NVIDIA Tesla A100).
For each case, both motion-compensated L1-SPIRiT and the proposed deep learning-based motion-compensated reconstruction were performed for comparison. An experienced cardiologist graded both reconstructions (5, excellent; 1, poor).
Results
Faster motion-corrected image reconstruction was demonstrated using the proposed deep learning-based framework. The reconstruction time was ~3 s per slice on an A100 GPU, while the reconstruction time of using (SMS-)L1-SPIRiT with 30 iterations on an Intel Xeon CPU (2.40 GHz) was ~45 minutes per slice. Furthermore, the image quality scores for using deep learning versus L1- SPIRiT reconstruction were 4.3 and 4.3 respectively. Figure 2 shows example slices from a patient undergoing clinical CMR perfusion imaging with an image quality of 5 using the deep learning-based image reconstruction. Figure 3 displays the deep learning-based rapid motion correction results, using our proposed technique, for the same Figure 2 patient case.Discussion and Conclusion
The proposed deep learning-based rapid motion-corrected image reconstruction framework enabled rapid and high-quality image reconstruction and motion correction for high-resolution spiral first-pass perfusion imaging at 3 T. The deep-learning motion correction framework provides more rapid motion correction than traditional off-line registration frameworks.Acknowledgements
This work was supported by NIH R01 HL131919.References
1. Tsao CW, Aday AW, Almarzooq ZI, Beaton AZ, Bittencourt MS, Boehme AK, et al. Heart Disease and Stroke Statistics—2022 Update: A Report From the American Heart Association. Circulation. 2022;145(8):e153–e639.
2. Gulati M, Levy PD, Mukherjee D, et al. 2021 AHA/ACC/ASE/CHEST/SAEM/SCCT/SCMR Guideline for the Evaluation and Diagnosis of Chest Pain: A Report of the American College of Cardiology/American Heart Association Joint Committee on Clinical Practice Guidelines. Circulation 2021;144:e368–e454.
3. Wang J, Yang Y, Weller DS, et al. High spatial resolution spiral first-pass myocardial perfusion imaging with whole-heart coverage at 3 T. Magnetic Resonance in Medicine 2021;86:648–662.
4. Wang J, Weller DS, Kramer CM, Salerno M. DEep learning-based rapid Spiral Image REconstruction (DESIRE) for high-resolution spiral first-pass myocardial perfusion imaging. NMR in Biomedicine 2022;35:e4661.
Figures
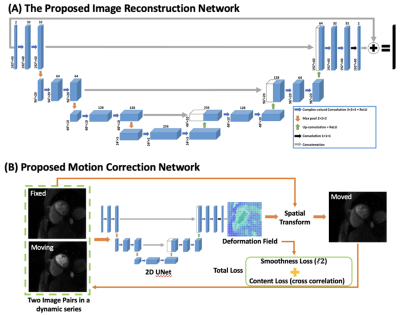
Figure 1. (A) The image reconstruction and (B) proposed image motion correction network for spiral perfusion imaging using deep learning.
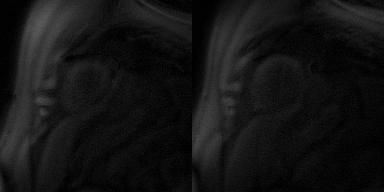
Figure 2. DESIRE reconstruction on a spiral perfusion imaging case, displaying two of the slices with image size 192 x 192 x 40 (frames).
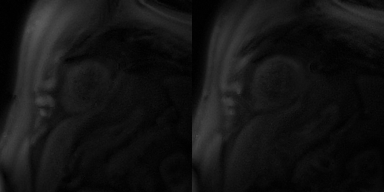
Figure 3. Motion correction performance of the slices in Figure 2 using the proposed deep learning-based technique.

Figure 4. Sample slice reconstruction from NUFFT. From left to right, NUFFT, L1-SPIRiT and DESIRE reconstructed slice.