0550
Highly Accelerated Cartesian Real-Time Cine CMR Using Subject-Specific Zero-Shot Deep Learning Reconstruction1Electrical and Computer Engineering, University of Minnesota, Minneapolis, MN, United States, 2Center for Magnetic Resonance Research, University of Minnesota, Minneapolis, MN, United States, 3Department of Radiology, Mayo Clinical, Rochester, MN, United States, 4National Heart-Lung and Blood Institute, Bethesda, MD, United States
Synopsis
Keywords: Heart, Heart
Real-time cine CMR is an ECG-free free-breathing alternative for functionally assessing the heart. To achieve sufficient spatio-temporal resolutions, these require rapid imaging, e.g. compressed sensing (CS) with radial trajectories. However, at high accelerations, CS may suffer from residual aliasing and temporal blurring. Recently, deep learning (DL) reconstruction has gained immense interest for fast MRI. Yet, for free-breathing real-time cine, where subjects have different breathing and cardiac motion patterns, database learning of spatiotemporal correlations has been difficult. Here, we propose a physics-guided DL reconstruction trained in a subject-specific manner. Proposed method improves image quality compared to database-trained DL and conventional methods.INTRODUCTION
Real-time cine CMR is an ECG-free free-breathing alternative to the clinical gold-standard ECG-gated, breath-hold cine MRI for evaluating cardiac function1. It is primarily used in patients who cannot hold their breaths or with arrhythmias, or during exercise stress2,3. In order to achieve high spatio-temporal resolution in real-time cine CMR, accelerated imaging, such as compressed sensing (CS), is often required, typically in conjunction with radial trajectories that exhibit incoherent artifacts, and flexibility with retrospectively adjusting temporal resolution4,5,6. Cartesian acquisitions with uniform undersampling are also used clinically, albeit at lower spatio-temporal resolutions7,8. Furthermore, CS reconstruction may suffer from residual aliasing and blurring at higher acceleration rates. Recently, deep learning (DL) reconstruction has shown promising results for highly-accelerated MRI, improving on parallel imaging and CS9,10. Yet, learning a spatiotemporally-regularized reconstruction for free-breathing real-time cine CMR, where subjects exhibit varying breathing and cardiac motion patterns, has been difficult. In this work, we use subject-specific zero-shot11 physics-guided DL (PG-DL) without a training database or ground-truth data to improve highly accelerated real-time cine CMR. Results on retrospectively 7.2-fold accelerated data show substantially improvement compared to existing methods.METHODS
Subject-Specific PG-DL Reconstruction:Regularized MRI reconstruction is formulated as:
$$\mathbf{x}=\arg \min _{\mathbf{x}}\left\|\mathbf{y}_{\Omega}-\mathbf{E}_{\Omega} \mathbf{x}\right\|_2^2+{\mathscr R}(\mathbf{x}), \quad\quad\quad (1)$$
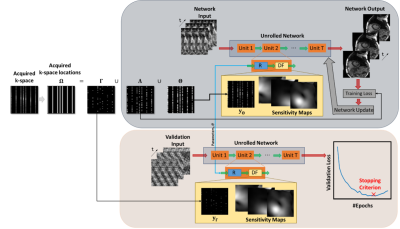
where $$$\mathbf{y}_{\Omega}$$$ is the acquired multi-coil k-space, Ω is the undersampling pattern,$$$\mathbf{E}_{\Omega}$$$ is the multi-coil encoding operator, $$$\mathbf{x}$$$ is the underlying image and $$${\mathscr R}$$$ is a regularizer. PG-DL methods solve this via algorithm unrolling12, alternating between conventional data-fidelity and a proximal operator, implicitly implemented via neural networks. While supervised learning with a fully-sampled reference is commonly used, recently self‐supervised learning, e.g. SSDU12, which does not require fully-sampled training data has been proposed. SSDU splits Ω into two disjoint sets $$$\Theta$$$ and $$$\Lambda$$$, where $$$\Theta$$$ enforces data-fidelity in the PG-DL network, while $$$\Lambda$$$ is used to define k-space loss. More recently, zero-shot self-supervised learning (ZS-SSDU)11 has been proposed for training a PG-DL network on a single undersampled dataset. In this approach, Ω is split into three disjoint sets as $$$\Omega = \Theta \cup \Lambda \cup \Gamma$$$ where $$$\Gamma$$$ is an additional set for defining a k-space self-validation loss for early stopping to avoid overfitting (Fig. 1)11. Using multi-mask data-augmentation13 that partitions $$$\Omega$$$ \ $$$\Gamma$$$ into K disjoint pairs $$$( \Omega_k,\Gamma_k)$$$, ZS-SSDU loss is given as:
$$\min _{\boldsymbol{\theta}} \frac{1}{K} \sum_{\mathrm{k}=1}^K {\mathscr L}\left(\mathbf{y}_{\Lambda_{\mathrm{k}^{\prime}}}, \mathbf{E}_{\Lambda_{\mathrm{k}}}\left({\mathscr f}\left(\mathrm{y}_{\Theta_{\mathrm{k}^{\prime}}} \mathbf{E}_{\Theta_{\mathrm{k}}} ; \boldsymbol{\theta}\right)\right)\right), \quad\quad\quad (2)$$
where $$$\boldsymbol{\theta}$$$ denotes the network parameters, $$${\mathscr f}(\cdot,\cdot)$$$ denotes the PG-DL network output and $$${\mathscr L}(\cdot,\cdot)$$$ is the training loss. This is supplemented with a self-validation loss on $$$\Gamma$$$ which is calculated at each epoch j from the current network weights specified by $$$\boldsymbol{\theta}^{(j)}$$$ as follows:
$${\mathscr L}\left(\mathbf{y}_{\Gamma}, \mathbf{E}_{\Gamma}\left({\mathscr f}\left(\mathbf{y}_{\Omega \backslash \Gamma}, \mathbf{E}_{\Omega \backslash \Gamma} ; \boldsymbol{\theta}^{(j)}\right)\right)\right). \quad\quad\quad (3)$$
When training on a single dataset, training loss in Eq. (2) keeps decreasing, and training is stopped once self-validation loss in Eq. (3) starts increasing to avoid overfitting11.
Imaging Experiments:
Real-time imaging datasets were acquired at 1.5T on 11 subjects with time-interleaved Cartesian acquisitions at acceleration R=4 using a bSSFP sequence. Relevant imaging parameters: resolution=2.25×2.93mm2, FOV=360×270mm2, temporal resolution=39ms and slice-thickness=8mm (12 slices). Real-time data were further retrospectively undersampled to R=7.2 by sampling every 8th line, while keeping the closest ky line to central k-space for each time-frame for a total of 10 lines per time-frame.
PG-DL Implementation Details:
Subject-specific regularization across all time-frames was performed using ZS-SSDU11 (Fig. 1) on a test subject. A 3D PG-DL network unrolled ADMM algorithm for 10 iterations. A 3D ResNet14 was used for the regularizer, with 50 time-frames in its input and 128 channels in hidden layers. Conjugate gradient was used for data fidelity15. Adam optimizer was used with LR=$$$2\cdot10^{-4}$$$ and mixed normalized $$$\ell_1-\ell_2$$$ loss12 were used. R adjacent frames were used to generate calibration data in a sliding-window manner16,17, on which coil sensitivity maps were generated18. 20% of $$$\Omega$$$ was uniformly randomly selected for $$$\Gamma$$$.
For R=7.2 data, comparisons were made to TGRAPPA15, locally-low-rank (LLR) regularized reconstruction19 and database-trained (10 subjects for training, 1 unseen subject for testing) SSDU12. TGRAPPA15 at standard acceleration R=4 was used for visual image quality comparisons.
RESULTS
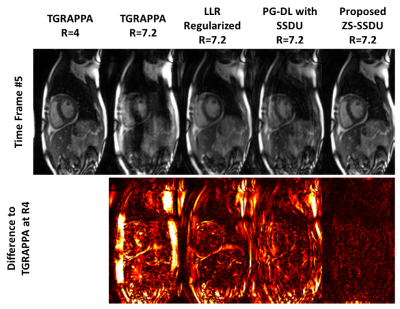
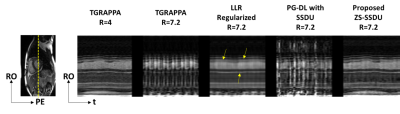
Fig. 2 shows an example time-frame reconstructed with different techniques. TGRAPPA and database-trained SSDU show substantial residual aliasing, former due to the high in-plane acceleration, and latter due to varying spatio-temporal correlations among subjects in the database. LLR-regularized reconstruction reduces residual aliasing albeit with temporal blurring. Proposed approach improves upon all methods and successfully suppresses aliasing. Difference images to TGRAPPA at R=4 show that proposed method only exhibits noise-like differences, whereas all other methods reveal structural differences.Fig. 3 depicts representative temporal intensity profiles. Temporal variations due to cardiac and respiratory motion are not preserved in TGRAPPA and SSDU, while LLR-regularized reconstruction exhibits temporal blurring. Proposed technique closely matches the R=4 data in terms of temporal intensity profiles.
DISCUSSION AND CONCLUSIONS
In this study, we proposed a subject-specific zero-shot PG-DL approach that does not require a training database or reference data for highly-accelerated real-time cine CMR, improving on existing methods. Use of zero-shot training enables regularization over spatio-temporal domain, where subject-specific breathing and cardiac motions are reliably learned without temporal blurring. Further studies with prospective higher accelerations at improved spatio-temporal resolutions are warranted.Acknowledgements
Funding: Grant support: NIH R01HL153146, NIH P41EB027061, NIH R21EB028369, NSF CCF-1651825.References
[1] Nayak, Krishna S., et al. "Real‐time cardiac MRI at 3 Tesla." Magnetic Resonance in Medicine: An Official Journal of the International Society for Magnetic Resonance in Medicine 51.4 (2004): 655-660.
[2] Zhang, Shuo, et al. "Real-time magnetic resonance imaging of cardiac function and flow—recent progress." Quantitative imaging in medicine and surgery 4.5 (2014): 313.
[3] Campbell‐Washburn, Adrienne E., et al. "Real‐time MRI guidance of cardiac interventions." Journal of Magnetic Resonance Imaging 46.4 (2017): 935-950.
[4] Haji‐Valizadeh, Hassan, et al. "Validation of highly accelerated real‐time cardiac cine MRI with radial k‐space sampling and compressed sensing in patients at 1.5 T and 3T." Magnetic resonance in medicine 79.5 (2018): 2745-2751.
[5] Uecker, Martin, Shuo Zhang, and Jens Frahm. "Nonlinear inverse reconstruction for real‐time MRI of the human heart using undersampled radial FLASH." Magnetic Resonance in Medicine: An Official Journal of the International Society for Magnetic Resonance in Medicine 63.6 (2010): 1456-1462.
[6] Feng, Li, et al. "XD‐GRASP: golden‐angle radial MRI with reconstruction of extra motion‐state dimensions using compressed sensing." Magnetic resonance in medicine 75.2 (2016): 775-788.
[7] Kellman, Peter, et al. "High spatial and temporal resolution cardiac cine MRI from retrospective reconstruction of data acquired in real time using motion correction and resorting." Magnetic Resonance in Medicine: An Official Journal of the International Society for Magnetic Resonance in Medicine 62.6 (2009): 1557-1564.
[8] Hansen, Michael S., et al. "Retrospective reconstruction of high temporal resolution cine images from real‐time MRI using iterative motion correction." Magnetic Resonance in Medicine 68.3 (2012): 741-750.
[9] K. Hammernik, T. Klatzer, et al., “Learning a variational network for reconstruction of accelerated MRI data,” Magn Reson Med, vol. 79, pp. 3055–3071, 2018.
[10] J. Schlemper, J. Caballero, J. V. Hajnal, A. N. Price, and D. Rueckert, “A deep cascade of convolutional neural networks for dynamic MR image reconstruction,” IEEE Trans Med Imaging, vol. 37, no. 2, pp. 491–503, 2017.
[11] Yaman, Burhaneddin, et al. "Zero-Shot Self-Supervised Learning for MRI Reconstruction." ICLR, 2022.
[12] Yaman, Burhaneddin, et al. "Self‐supervised learning of physics‐guided reconstruction neural networks without fully sampled reference data." Magnetic Resonance in Medicine 84.6 (2020): 3172-3191.
[13] Yaman, Burhaneddin, et al. "Multi‐mask self‐supervised learning for physics‐guided neural networks in highly accelerated magnetic resonance imaging." NMR in Biomedicine (2022): e4798.
[14] Timofte, Radu, et al. "NTIRE 2017 Challenge on Single Image Super-Resolution: Methods and Results.” IEEE Computer Society Conference on Computer Vision and Pattern Recognition Workshops; 2017 Jul 21-26; Honolulu, HI. p 1110-1121.
[15] Aggarwal HK, Mani MP, Jacob M. MoDL: Model-based deep learning architecture for inverse problems. IEEE Trans Med Imaging. 2019; 38: 394- 405.
[16] Breuer, Felix A., et al. "Dynamic autocalibrated parallel imaging using temporal GRAPPA (TGRAPPA)." Magnetic Resonance in Medicine: An Official Journal of the International Society for Magnetic Resonance in Medicine 53.4 (2005): 981-985.
[17] Kellman, Peter, Frederick H. Epstein, and Elliot R. McVeigh. "Adaptive sensitivity encoding incorporating temporal filtering (TSENSE)." Magnetic Resonance in Medicine,45.5 (2001): 846-852.
[18] M. Uecker, P. Lai, et al., “ESPIRiT—an eigenvalue approach to autocalibrating parallel MRI: where SENSE meets GRAPPA,” Magn Reson Med, vol. 71, no. 3, pp. 990–1001, 2014.
[19] Miao, Xin, et al. "Accelerated cardiac cine MRI using locally low rank and finite difference constraints." Magnetic resonance imaging 34.6 (2016): 707-714.
Figures