0525
Differentiation of recurrent tumor from post-treatment changes in Glioblastoma patients using Deep Learning and Restriction Spectrum Imaging1Department of Radiology, University of California San Diego, La Jolla, CA, United States, 2Cortechs.ai, San Diego, CA, United States, 3Department of Pathology, University of California San Diego, La Jolla, CA, United States, 4Department of Radiation Medicine and Applied Sciences, University of California San Diego, La Jolla, CA, United States, 5Department of Neurological Surgery, University of California San Diego, La Jolla, CA, United States, 6Department of Bioengineering, University of California San Diego, La Jolla, CA, United States, 7Department of Translational Neurosciences, Pacific Neuroscience Institute and Saint John’s Cancer Institute at Providence Saint Johns’ Health Center, Santa Monica, CA, United States, 8Department of Neuroscience, University of California San Diego, La Jolla, CA, United States
Synopsis
Keywords: Tumors, Diffusion/other diffusion imaging techniques
Differentiating recurrent tumor from post-treatment changes is challenging in glioblastoma. Using restriction spectrum imaging (RSI) and deep learning, we were able to accurately identify and segment residual and recurrent enhancing and non-enhancing cellular tumor in post-treatment brain MRIs. Including RSI in the deep learning model improved tumor segmentation due to the ability of RSI to separate cellular tumor from peritumoral edema and treatment related enhancement. The volume of cellular tumor was also predictive of survival. Our results suggest that combining deep learning and RSI may identify recurrent tumor in glioblastoma patients, which could improve targeted treatments and guide clinical decision-making.
Introduction
Differentiating recurrent tumor from post-treatment changes induced by the combination of chemotherapy, radiotherapy and anti-angiogenic agents represents a challenge in post-operative brain MRI studies of glioblastoma patients1,2,3. This problem is crucial in the management of these patients since a delay in the diagnosis of recurrent tumor may prevent the patient from receiving prompt treatment, while overcalling recurrent tumor may result in unnecessary surgery and/or change in therapy.Two major features account for the difficulty of differentiating these entities: 1) both post-treatment changes and recurrent tumor can enhance and 2) both infiltrative tumor and peritumoral edema can result in increased T2 signal intensity. To address this problem, we used a synergistic combination of recent state-of-the-art technologies: 1) a multi-shell diffusion weighted MR sequence termed Restriction Spectrum Imaging (RSI) specifically designed to separate infiltrative tumor from edema4,5 and 2) the nnU-Net, which is a powerful deep learning convolutional neural network, that has excelled in the segmentation of brain tumors in multimodal MRI volumes6,7.
Methods
A multimodal MRI data set including T1 pre and post contrast, FLAIR, DWI, ADC and RSI-derived maps of restricted isotropic diffusion (“RSI cellularity”) in patients with glioblastoma was used. A total of 88 MRIs from 62 patients were manually segmented for cellular tumor by an attending neuroradiologist, which included 68 cases with recurrent enhancing or non-enhancing cellular tumor and 20 cases with post treatment changes. Of the 88 cases, 9 were either pre-operative or post biopsy, with the remaining performed after surgical resection. The Deep Learning, 3D nnU-Net model was trained using 68 randomly selected volumes and tested on the remaining 20 unique volumes.Results
An example of segmentation for a cellular non-enhancing tumor case is shown in Fig 1. While the regions of FLAIR hyperintensity in the splenium of the corpus callosum and bilateral parieto-occipital white matter did not demonstrate significant enhancement, they showed increased RSI cellularity signal compatible with cellular infiltrative tumor (Fig 1A). These regions were segmented as tumor tissue in Fig 1B. An example of segmentation for a case with only post-treatment changes is shown in Fig. 2. The red arrow in Fig 2A demonstrated peripheral nodular enhancement at the anterior margin of the surgical cavity but the RSI cellularity map (blue arrow) did not demonstrate RSI cellularity signal in the enhancing regions or surrounding edematous tissue, suggesting post-treatment changes rather than recurrent tumor. This was confirmed on the follow-up scan (Fig 2B) where this region of nodular enhancement improved (green arrow) and tissue necrosis increased (yellow arrow) as shown by the very high RSI signal. Tumor segmentations with several variations of the Deep Learning model with different inputs are shown in Fig 3 for an individual test subject and the average Dice scores over the entire testing set is shown in Table 1. Adding RSI cellularity to the combination of T1 pre, T1C+ and FLAIR sequences increased the Dice score from 0.50 to 0.65. Although adding ADC to T1, T1C+, and FLAIR sequences also increased the Dice score, the increase was less pronounced than with RSI (0.60 ± 0.31 vs. 0.65 ± 0.24)." Fig 4 demonstrates an inverse relationship between volume of enhancing cellular tumor (both T1C+ and RSI positive) and the overall survival for patients with RSI scans within 180 days following surgical resection (n=20).Discussion
Our results demonstrate that adding either ADC or RSI improves accuracy of the deep learning model to segment tumor on the post-operative MRI scan, reflecting the ability of diffusion-weighted imaging to provide information about tissue content independent of contrast enhancement, which is a measure of the blood brain barrier permeability. Furthermore, the improvement in Dice score was greater using RSI cellularity compared to ADC, as RSI provides a more specific measure of spherically restricted diffusion compared to standard DWI, which is contaminated by hindered diffusion processes such as peritumoral edema.While the current model considers both tumor cellularity (via RSI) as well as blood brain barrier permeability (via T1C+), our future work will also include perfusion measurements and susceptibility weighted imaging (SWI). We anticipate that including these additional sequences in our Deep Learning model will further improve tumor segmentation and result in higher Dice scores, in addition to increasing sample size.
Our results also demonstrate that the volume of enhancing cellular tumor (both T1C+ and RSI positive) is a potential biomarker of overall survival. We are currently collecting more data to better characterize the relationship between enhancing cellular tumor and survival. Moreover, we expect that including additional MRI sequences such as perfusion and SWI (as described above) will result in a more accurate biomarker to predict patient survival from post-operative MRI scans.
Conclusions
In this work, we showed that including DWI MRI in a deep learning model improved tumor segmentation in post-operative MRI scans. The model performed better using RSI-derived imaging biomarkers of cellularity compared with traditional ADC. Our results are promising and suggest that combining deep learning and RSI may differentiate recurrent tumor from post treatment changes in glioblastoma patients and predict survival, which is a major challenge in neuro-oncology. Future work will also include perfusion measurement and SWI to further improve the deep learning model.Acknowledgements
This research was funded by an NIH SBIR grant.
References
1. Rudie, J. D., Rauschecker, A. M., Bryan, R. N., Davatzikos, C. & Mohan, S. Emerging Applications of Artificial Intelligence in Neuro-Oncology. Radiology 290, 607–618 (2019).
2. Gilbert, M. R. et al. Dose-Dense Temozolomide for Newly Diagnosed Glioblastoma: A Randomized Phase III Clinical Trial. J Clin Oncol 31, 4085–4091 (2013).
3. Bray, F. et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. Ca Cancer J Clin 68, 394–424 (2018).
4. White, N. S., Leergaard, T. B., D’Arceuil, H., Bjaalie, J. G. & Dale, A. M. Probing tissue microstructure with restriction spectrum imaging: Histological and theoretical validation. Hum Brain Mapp 34, 327–346 (2013).
5. Kothari, P. D. et al. Longitudinal Restriction Spectrum Imaging Is Resistant to Pseudoresponse in Patients with High-Grade Gliomas Treated with Bevacizumab. Am J Neuroradiol 34, 1752–1757 (2013).
6. Isensee, F., Jaeger, P. F., Kohl, S. A. A., Petersen, J. & Maier-Hein, K. H. nnU-Net: a self-configuring method for deep learning-based biomedical image segmentation. Nat Methods 18, 203–211 (2021).
7. Rudie, J. D. et al. Longitudinal Assessment of Posttreatment Diffuse Glioma Tissue Volumes with Three-dimensional Convolutional Neural Networks. Radiology Artif Intell 4, e210243 (2022).
Figures
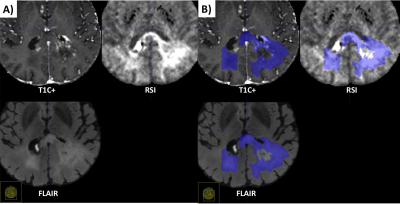
Fig. 1. Example of segmentation for a cellular non-enhancing tumor case. A) MRI volumes. While the hyper FLAIR signal regions in the splenium of the corpus callosum and bilateral parieto-occipital white matter did not demonstrated enhancement, they showed increased RSI signal demonstrating residual tumor. B) These regions were included in manual segmentations of cellular tumor.
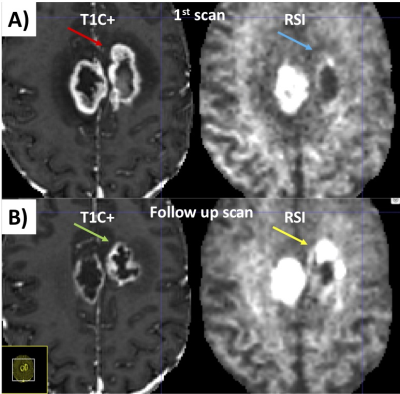
Fig. 2. Example case of a pure post-treatment changes. A) Initial post-surgical study. The red arrow demonstrates peripheral nodular enhancement at the anterior margin of the surgical cavity but the same region on the RSI map (blue arrow) does not demonstrate high cellularity, suggesting post-treatment changes rather than recurrent tumor. B) On the follow-up study, this region of nodular enhancement has resolved (green arrow) and tissue necrosis is now present (yellow arrow) as shown by the very high RSI signal.
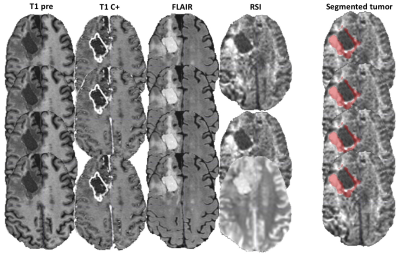
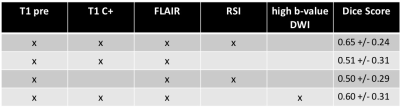
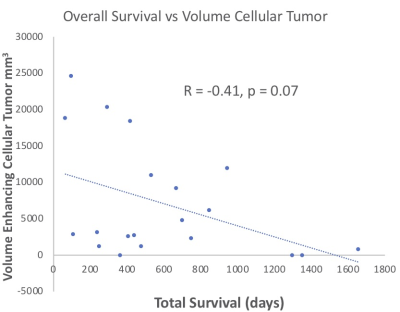
Fig. 4 Preliminary data for the use of RSI as a biomarker for survival. The volume of enhancing cellular tumor (positive enhancement and high signal on RSI) as a function of the total survival days. The dots represent patients (n=20) for which an MRI with the RSI sequence was performed in the first 180 days following the gross total resection of their glioblastoma.