0522
The MRI-based 3D-ResNet-101 deep learning model for predicting preoperative grading of gliomas: a multicenter study1Department of Magnetic Resonance, Lanzhou University Second Hospital, Lanzhou, China, 2Philips Healthcare, Xi'an, China
Synopsis
Keywords: Tumors, Machine Learning/Artificial Intelligence, Deep learning
The preoperative accurate and non-invasive prediction of glioma grading remains challenging. To accurately predict high-or low-grade gliomas, we constructed a 3D-ResNet101 deep learning model with data from a multicenter. These data were obtained from the Second Hospital of Lanzhou University, with 708 glioma patients, and the TCIA database, with 211 patients. The areas under the curve of the 3D-ResNet-101 deep learning model are 0.97 and 0.89 in the test cohort and external test cohort, respectively. This new method can be used for non-invasive prediction of glioma grading before surgery.Introduction
Glioma is the most common primary intracranial malignant tumor in adults with high disability and recurrence rates. Currently, glioma treatment is mainly surgical resection, followed by radiotherapy and chemotherapy. The surgical treatment principle of glioma is to maximize safe resection, but the surgical strategy of high-and low-grade glioma is slightly different. Patients with high-grade gliomas are older and more invasive, so the focus is on relieving intracranial hypertension based on maximum safe resection. Low-grade gliomas need to consider tumor location and comprehensive function protection1. Therefore, the strategy of surgical treatment still relies on the preoperative grading of the tumor. However, with the development of new MRI technology, the dramatic increase in image data has brought great difficulties and challenges to traditional image diagnosis. In recent years, the application of artificial intelligence in medical image processing has become a hot topic in the medical field. Deep learning is a computer-assisted methodology that has been proven that it can potentially predict glioma types in a non-invasive way. However, its classification performance deteriorates as deep learning optimizes deep neural networks. A residual system like ResNet has been reported to achieve higher accuracy in deeper neural networks and enables better representation of the neural network2. Nevertheless, most previous research is based on 2D ResNet, and it cannot fully exploit the 3D spatial contextual information of volumetric image data. Therefore, we attempted to construct a 3D-ResNet101 deep-learning model to predict glioma grading.Method
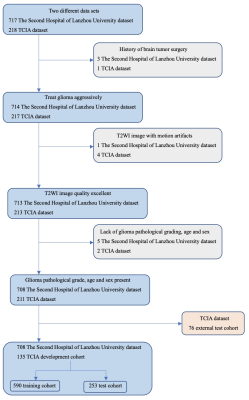
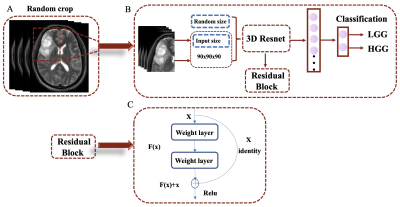
We retrospectively included 919 patients with glioma from the Second Hospital of Lanzhou University (n=708) and the TCIA database (n=211). The data set from TCAI is split into a development cohort (n=135) and an external test cohort (n=76). Figure 1 is a flowchart of patient enrollment. The patients collected at the Second Hospital of Lanzhou University are scanned by a 3.0T MR (Ingenia CX, Philips Healthcare, the Netherlands) with a 16-channel head coil. The following T2WI sequence parameters are TR 4500ms, TE 91ms; flip angle: 176°, matrix: 448×448, slice thickness: 3mm, slice spacing: 0.6mm, FOV: 230mm×230mm. The patients picked up from TCIA are scanned by a 1.5T MR (General Electric) with an 8-channel head coil, and the following T2WI sequence parameters are TR 4000ms, TE 85ms; flip angle: 156°, matrix: 448×448, slice thickness: 5mm, slice spacing: 0.5mm, FOV: 230mm×230mm. All images were downloaded anonymously in DICOM format. First, all data were N4 bias field corrected and then the 3D cubic patches (including tumor parenchyma and edema areas) were cropped and resampled to 90x90x90 size using the nearest neighbor method. Finally, the images were normalized and saved in the NIfTI format. A total of 843 cases from the Second Hospital of Lanzhou University and the TCIA development cohort were randomly divided into a training cohort (n=590) and a test cohort (n=253) with a ratio of 7:3. A 3D-ResNet101 deep learning models were constructed based on T2WI images; The training models are tested in a test queue and an external test queue. Furthermore, the model composition is shown in Figure 2. We used A one-way analysis of variance (ANOVA) to compare differences between patient age groups, and the gender data were analyzed using the chi-square test. P<0.05 is set to be statistically significant.Result
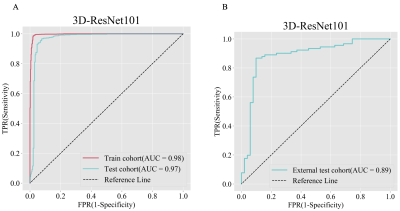
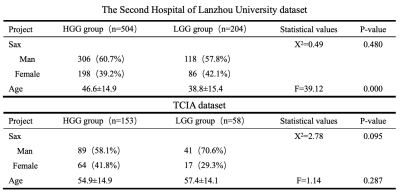
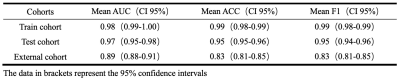
The clinical information of the patients is shown in Table 1. The 3D-ResNet101 deep learning model constructed based on T2WI images had an accuracy of 0.99 and 0.95 in the training and test cohort, respectively, with F1 scores of 0.99 and 0.95 and AUCs of 0.98 and 0.97; the external test cohort had an accuracy of 0.83, F1 score of 0.83 and AUC of 0.89 (Table 2 and Figure 3).Discussion
Our model is based on a T2WI sequence. In addition, the input images of the model are randomly cropped according to the tumor contours. This helps to extract tumor features while dropping redundant features and optimizes the depth and performance of the model. ResNet is currently one of the most popular deep learning architectures, and some previous studies have used deep learning models based on the ResNet architecture for tumor segmentation or differential diagnosis of gliomas3,4. The ResNet architecture is also used in our study but differs in that we utilize 3D CNN, which can fully use the 3D spatial contextual information of the volumetric image data. One study proposed 2D mask R-CNN and 3DconvNet methods for grading gliomas, respectively. Their results confirmed that the 3D-CNN approach performed better than the former one5. One researcher found that when using CNNs for brain tumor segmentation, the performance of tumor segmentation is significantly decreased when using data from different institutions for training compared to data from the same institution6. Our data come from different institutions. In the external test cohort, we tested the model and got good results, which further verified the robustness and repeatability of our model.Conclusion
Our results indicate that the T2WI-based 3D-ResNet101 deep learning model for differentiating between high- and low-grade gliomas is highly accurate and offers potential guarantees for further entry into clinical practice in this field.Acknowledgements
This work was supported by the Second Hospital of Lanzhou University - Cuiying Science and Technology Innovation (Top-notch Clinical Technology Research) (CY2021-BJ-A05)References
1. Jiang T, Nam DH, Ram Z, et al. Clinical practice guidelines for the management of adult diffuse gliomas [J]. Cancer Lett, 2021,499: 60-72.
2. He K, Zhang X, Ren S, et al. Deep Residual Learning for Image Recognition [J]. 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), 2016: 770-778.
3. Shin I, Kim H, Ahn SS, et al. Development and Validation of a Deep Learning-Based Model to Distinguish Glioblastoma from Solitary Brain Metastasis Using Conventional MR Images [J]. AJNR Am J Neuroradiol, 2021,42(5): 838-844.
4. Artzi M, Gershov S, Ben-Sira L, et al. Automatic segmentation, classification, and follow-up of optic pathway gliomas using deep learning and fuzzy c-means clustering based on MRI [J]. Med Phys, 2020,47(11): 5693-5701.
5. Zhuge Y, Ning H, Mathen P, et al. Automated glioma grading on conventional MRI images using deep convolutional neural networks [J]. Med Phys, 2020,47(7): 3044-3053.
6. AlBadawy EA, Saha A, Mazurowski MA. Deep learning for segmentation of brain tumors: Impact of cross-institutional training and testing [J]. Med Phys, 2018,45(3): 1150-1158.
Figures