0521
A Pretrained CNN Model Using Multiparametric MRI to Identify WHO Tumor Grade of Meningiomas1Institute of Biomedical Engineering, Bogazici University, Istanbul, Turkey, 2Department of Radiology, Basaksehir Cam and Sakura City Hospital, Istanbul, Turkey, 3Electric and Electronic Engineering Department, Bogazici University, Istanbul, Turkey, 4Brain Tumor Research Group, Acibadem University, Istanbul, Turkey, 5Center for Neuroradiological Applications and Reseach, Acibadem University, Istanbul, Turkey, 6Department of Medical Pathology, Acibadem University, Istanbul, Turkey, 7Department of Neurosurgery, Acibadem University, Istanbul, Turkey, 8Department of Radiology, Acibadem University, Istanbul, Turkey
Synopsis
Keywords: Tumors, Machine Learning/Artificial Intelligence
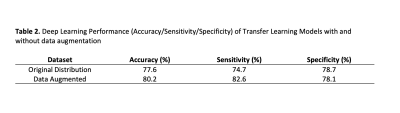
Meningiomas are the most common primary extra-axial intracranial tumors in adults. Grade of meningioma helps to predict the patient prognosis. Sixty-two patients with preoperative MRI were included in this IRB approved study. The whole tumor volumes were segmented from FLAIR, followed by co-registration onto SWI. A pretrained convolutional neural network (CNN) was employed to classify meningiomas into high and low-grades based on SWI, CE-T1W and FLAIR MRI. The pretrained CNN with data augmentation resulted in an accuracy of 80.2% (sensitivity=82.6% and specificity=78.1%) for identifying grades in meningiomas.Introduction
Meningiomas are mainly benign, slow-growing neoplasms that arise from two types of cells which are arachnoid and dural border cells (1). According to the updated World Health Organization (WHO) 2021 brain tumor classification, meningiomas are classified into three grades based on clinical and molecular genetic markers (2). Grade 1 (low-grade) meningioma accounts for over 90% of cases, while grades 2 and 3 (high grades) meningiomas are known to have aggressive behavior, resulting in significant mortality and morbidity. Low-grade meningiomas have a more prolonged progression-free survival than high-grade tumors (3, 4). When a mass lesion with radiological signs suggestive of meningioma is diagnosed, no reliable radiological markers could predict tumor grade. Therefore, non-invasive predictors of meningioma grade may be beneficial to clinical decision-making. Susceptibility weighted MRI (SWI) is sensitive to neovascularization and microcalcification. The ability of tumors to develop angiogenesis is a consequence of hypoxia. It is considered one of the critical features of tumor progression, enabling sustained growth and a change in the invasion characteristics, which are related to tumor grade (5). This study proposes a workflow incorporating SWI MRI and conventional MRI sequences to assess the relationship between imaging features and WHO grades in meningiomas using deep learning.Material-Methods
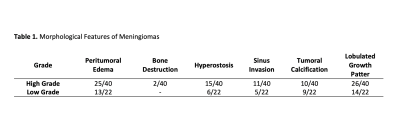
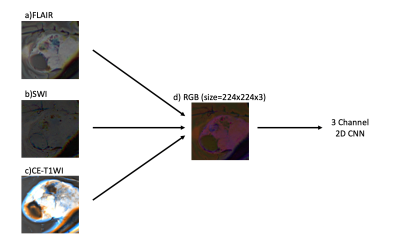
Sixty-two patients diagnosed with meningiomas (24M/38F), who underwent a preoperative MRI, were retrospectively recruited in this IRB-approved study. A senior pathologist examined tumor samples to assign grades according to the WHO classification system. MRI protocol included FLAIR (Time of repetition (TR)/Time of Echo (TE)=8320/92ms, TI =2000ms, field of view (FOV) =220mm, slice thickness=3mm), SWI (TR/TE=28/20ms, FOV=220mm, slice thickness=1.6mm), and Contrast-Enhanced-T1W MRI (CE-T1W) (TR/TE=589/10ms, FOV=220mm, slice thickness=3mm) on a 3T clinical MR scanner (Siemens Healthcare, Erlangen, Germany). Morphological features, including sinus invasion, bone destruction, hyperostosis, peritumoral edema, growth pattern, and tumoral calcification were compared among the low and high grade groups with chi-square or Ficher’s exact test. 3D Slicer version 4.8.1 (http://slicer.org/) was used to segment the tumor volumes on FLAIR. The CE-T1W and FLAIR images were registered to the high-resolution SWI. The transformation matrix (FSL; http://fsl.fmrib.ox.ac.uk/fsl/fslwiki/) was used to register the segmentation masks for each subject onto the SWI. The longest segmentation in both x and y directions determined the slice of each modality and the rectangular bounding box around the lesion. Cropped area of the selected slice on each modality was merged to create red, green, and blue (RGB) images using MATLAB (MathWorks Inc., Natick, MA, USA). RGB image inputs were resized to 224x224x3. Additional data were generated via an image data generator in Keras to overcome the data imbalance problem. Scaler standardization was utilized for preprocessing. An EfficientNetV2 with pre-trained weights was used for training, with only changing the densely connected layers (6). The learning rate and batch size were set to 8x10-6 and 4, respectively. The model was trained twice with and without data augmentation. In the validation cohort, binary cross-entropy loss function and accuracy were utilized to assess the model performance.Results
Forty meningiomas were high grade (WHO grades 2 and 3), and 22 were low grade. Morphological features of the patient cohort are summarized in Table 1. Radiological morphological features were similar between the two groups (P>0.05). The prediction performance of the transfer learning (EfficientNetv2) method with data augmentation was higher than the pre-trained model with the original distributed dataset (Table 2). While the transfer learning approach with data augmentation had a higher accuracy of 80.2% (sensitivity=82.6%, specificity=78.1%), the transfer learning model trained with the original distributed dataset resulted in an accuracy of only 77.6 % (sensitivity=74.7 %, specificity=78.7 %) for predicting grade in this small patient cohort (Table 2).Discussion-Conclusion
The outcomes of this study indicated that deep learning models could predict grades of meningioma using fused SWI-MRI, CE-T1W and FLAIR images. Semantic features depending on visual assessment were similar between high and low-grade meningiomas in this dataset. The proposed deep learning model identified the grade in meningiomas with 80% accuracy using hierarchical convolution operations of the image. In addition, high-order features beyond the visual assessment were extracted from SWI-MRI, CE-T1W images, and FLAIR using the pre-trained network. Future studies will validate deep learning results in a larger patient cohort.Acknowledgements
This study was supported by the Scientific and Technological Research Council of Turkey (TUBITAK) grant 119S520.References
1. Yamashima T. Human Meninges: Anatomy and Its Role in Meningioma Pathogenesis. In: Lee JH, editor. Meningiomas. London: Springer London; 2009. p. 15-24.
2. Louis DN, Perry A, Wesseling P, Brat DJ, Cree IA, Figarella-Branger D, et al. The 2021 WHO Classification of Tumors of the Central Nervous System: a summary. Neuro Oncol. 2021;23(8):1231-51.
3. Champeaux C, Wilson E, Shieff C, Khan AA, Thorne L. WHO grade II meningioma: a retrospective study for outcome and prognostic factor assessment. J Neurooncol. 2016;129(2):337-45.
4. Nabors LB, Portnow J, Ammirati M, Baehring J, Brem H, Butowski N, et al. NCCN Guidelines Insights: Central Nervous System Cancers, Version 1.2017. J Natl Compr Canc Netw. 2017;15(11):1331-45.
5. Niu G, Chen X. Vascular endothelial growth factor as an anti-angiogenic target for cancer therapy. Curr Drug Targets. 2010;11(8):1000-17.
6. Tan ML, Q.V. EfficientNetV2: Smaller Models and Faster Training. arXiv 2021.arXiv:2104.00298.
Figures