0491
sLASER Performed Similarly to PRESS at Revealing Metabolite-Age Correlations1Radiology and Radiological Sciences, Johns Hopkins University School of Medicine, Baltimore, MD, United States, 2F.M. Kirby Research Center for Functional Brain Imaging, Kennedy Krieger Institute, Baltimore, MD, United States, 3Department of Radiology, Shandong Provincial Hospital Affiliated to Shandong First Medical University, Jinan, China, 4Department of Radiology, Shandong University, Jinan, China
Synopsis
Keywords: Spectroscopy, Data Analysis, magnetic resonance spectroscopy, sLASER, PRESS, localization, aging
Recent MRS community consensus recommended sLASER over PRESS for reduced chemical-shift displacement. There is very little evidence supporting this consensus in terms of ability to reveal in vivo biochemistry. sLASER- and PRESS-localized spectra were collected in gray- and white-matter regions in 102 adult subjects (aged 20-69). sLASER showed slightly higher SNR than PRESS (by 4% on average), but improved SNR and localization did not convert into reduced variance or improved detection of metabolite-age correlations. Between-subject CVs of 13 modeled metabolites were remarkably consistent, and the pattern of metabolite-age correlations was also similar.Introduction
Recent single-voxel MRS community consensus (1,2) recommended sLASER (3) over PRESS (4), a method that has been widely applied for over two decades. This recommendation is motivated by improved localization performance, in particular reduced chemical-shift displacement artefact (CSDA; since sLASER uses inversion pulses with higher bandwidth than the refocusing pulses of PRESS). While improved localization is desirable, it is not the single factor limiting single-voxel MRS, and it is important not only to demonstrate that sLASER is theoretically preferable, but also that the newer methodology has improved power to interrogate in vivo biochemistry.There is an extensive literature of age-related changes in metabolite levels measured with MRS (5,6), making age an appropriate external validation variable to compare the performance of sLASER and PRESS. If sLASER is a better methodology for applied MRS studies of the brain that PRESS, we would expect it to show: increased SNR and reduced linewidth (due to improved localization); decreased variance in metabolite measurements made with sLASER; and increased sensitivity to metabolite-age correlations.
Methods
A sex- and age-balanced cohort of 102 healthy volunteers was recruited with local IRB approval (53 female; aged 20-69). A Philips 3T MRI scanner with a 32-channel phased-array head coil for RF receive was used to acquire MRS data with sLASER and PRESS localization in two voxels localized in the centrum semiovale (CSO) and the posterior cingulate cortex (PCC) regions (30 × 26 × 26 mm3). T1-weighted MPRAGE (TR/TE/ 6.9/3.2 ms; FA 8°; 1 mm3 isotropic resolution) was acquired for voxel positioning and tissue segmentation. PRESS- (refocusing: bandwidth 1.3 kHz; duration 6.90 ms) and sLASER- (inversion: bandwidth 10 kHz; duration 4.5 ms) localized data were acquired with: TR/TE 2000/30 ms; 96 transients of 2048 datapoints sampled at 2 kHz; Philips-CHESS water suppression (bandwidth 140 Hz). A 20-mm slice-selective saturation pulse was applied to suppress subcutaneous lipid adjacent to the voxel. Water reference spectra were also acquired.Spectra were processed and modeled using the Osprey software (7), following consensus methods (8). PRESS- and sLASER-basis sets consisting of 18 simulated metabolite basis functions and macromolecules were employed. Tissue segmentation was performed in SPM12 (9). Signal-to-noise ratio (SNR), full-width-half-maximum linewidth of NAA, and metabolite concentrations were extracted.
All statistical analyses were performed using R (Version 4.0.2). One-way ANOVA were used to compare SNR and linewidth of NAA. Coefficients of variation (CV) were calculated for each metabolite measurements. Pearson correlation coefficient (r) were used to test the relationship between PRESS and sLASER datasets and their correlations to the metabolite age profiles. P-values less than 0.05 were considered statistically significant.
Results
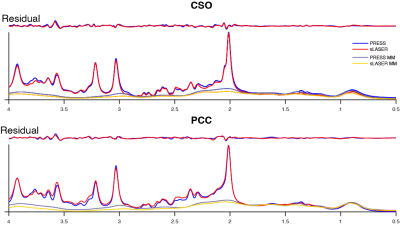
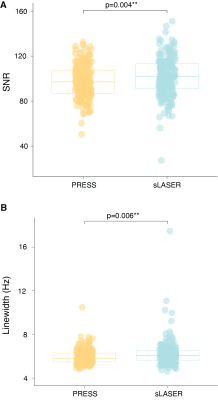
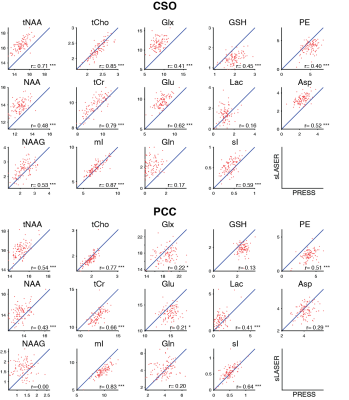
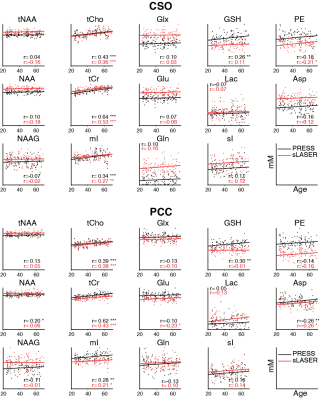
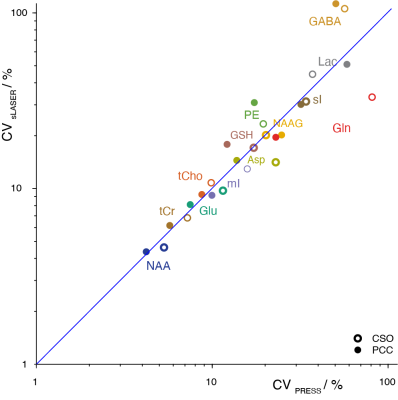
Mean spectra acquired from 102 volunteers are shown in Figure 1. Two PRESS and two sLASER spectra were excluded due to lipid contamination. SNR was 4% higher (p<0.05) on average for sLASER than PRESS (Figure 2A). Linewidth was also 4% higher (p<0.05) on average for sLASER (Figure 2B). PRESS and sLASER metabolite measures were significantly correlated (p<0.05 for all modeled metabolites except Lac and Gln in CSO and GSH, Gln and NAAG in PCC; Figure 3), but with modest correlation coefficients (up to 0.8 for tCho and mI). Significant metabolite-age relationships were observed for both PRESS and sLASER for tCho, tCr, and mI in both regions and for Asp in PCC (Figure 4). In more cases the correlation coefficient was higher for PRESS. PRESS data suggested age-related increases in GSH that sLASER did not (in both regions), and for NAA in PCC. Most metabolites of interest showed similar CVs for PRESS and sLASER (ranging from ~5% for NAA to over 30% for Lac, Gln and GABA; Figure 5).Discussion
Overall, there is very little in this dataset to differentiate our implementation of PRESS and sLASER localization. Spectral quality, including modeling residuals, are very similar. sLASER give slightly improved SNR, most likely due to improve within-slice signal. The reduced CSDA of sLASER did not yield improved NAA linewidth, presumably because the increased localization bandwidth did not give substantially better shim in the NAA-excited region (shifted by 10% from the prescribed voxel for PRESS and by 1.3% for sLASER). If anything, sLASER may have given worse linewidth in PCC (posthoc t-test p<0.05). PRESS and sLASER measurements were well-correlated for tNAA, tCho, tCr, mI and sI, and more modestly correlated for the other metabolites. Even metabolites for which simple linear combination modeling of unedited spectra is questionable showed significant between-method correlations (e.g. GSH and PE in CSO). Where agreement between methods was surprisingly poor (e.g. in Glx), this could not be explained by differences in method performance, since within-method variance in metabolite measures was similar.Relative to age, which is treated in this study as an external validation measure, there is no evidence to support the hypothesis that sLASER is better at revealing biological metabolite relationships than PRESS. If anything, the age-metabolite correlation coefficients sightly favor PRESS. This is an important result, because current community consensus favors replacing a method that has been extremely widely tested in favor of one that is relatively poorly understood, on the basis not of experimental evidence of improved ability to learn about the brain, but a rather obscure technical detail.
Acknowledgements
This work was supported by NIH grants R01 EB016089, R01 EB023963, R21 AG060245, R00 AG062230, K99 DA051315 and P41 EB031771.References
1. Wilson M, Andronesi O, Barker PB, et al. Methodological consensus on clinical proton MRS of the brain: Review and recommendations. Magn Reson Med 2019;82(2):527-550.
2. Oz G, Deelchand DK, Wijnen JP, et al. Advanced single voxel (1) H magnetic resonance spectroscopy techniques in humans: Experts' consensus recommendations. NMR Biomed 2020:e4236.
3. Scheenen TW, Heerschap A, Klomp DW. Towards 1H-MRSI of the human brain at 7T with slice-selective adiabatic refocusing pulses. MAGMA 2008;21(1-2):95-101.
4. Bottomley PA. Spatial localization in NMR spectroscopy in vivo. Ann N Y Acad Sci 1987;508:333-348.
5. Bozgeyik Z, Burakgazi G, Sen Y, Ogur E. Age-related metabolic changes in the corpus callosum: assessment with MR spectroscopy. Diagn Interv Radiol 2008;14(4):173-176.
6. Brandt AS, Unschuld PG, Pradhan S, et al. Age-related changes in anterior cingulate cortex glutamate in schizophrenia: A (1)H MRS Study at 7 Tesla. Schizophr Res 2016;172(1-3):101-105.
7. Oeltzschner G, Zollner HJ, Hui SCN, et al. Osprey: Open-source processing, reconstruction & estimation of magnetic resonance spectroscopy data. J Neurosci Methods 2020;343:108827.
8. Near J, Harris AD, Juchem C, et al. Preprocessing, analysis and quantification in single-voxel magnetic resonance spectroscopy: experts' consensus recommendations. NMR Biomed 2021;34(5):e4257.
9. Friston KJ, Holmes AP, Worsley KJ, Poline JP, Frith CD, Frackowiak RSJ. Statistical parametric maps in functional imaging: A general linear approach. Human Brain Mapping 1994;2(4):189-210.
Figures