0483
Optimized fMRI preprocessing pipeline enables robust functional connectivity analysis of mouse brain at laminar level1University of Minnesota, Minneapolis, MN, United States
Synopsis
Keywords: Data Processing, fMRI
High-resolution BOLD fMRI has become an essential tool for studying neural circuit and hemodynamic changes at mesoscopic scale. Nevertheless, it is more prone to the poor sensitivity and non-neural signal contamination as the spatial resolution increases. Group-level analysis also imposes new requirements on the subject alignment accuracy. To deal with these challenges, we developed a fMRI preprocessing pipeline featured in random matrix theory-based PCA denoising, one-time image voxel shift correction, and enhanced subject-level alignment. We applied this pipeline to the high-resolution mouse resting state fMRI and achieved high-quality hierarchical connectomes from large brain regions to thin cortical layers.
Introduction
Highly correlated blood oxygenation level-dependent (BOLD)-based resting state functional magnetic resonance imaging (rs-fMRI) signals reflect the intrinsic connectivity among brain regions. The BOLD signals, nevertheless, are usually weak and contaminated by thermal noise, motion artifacts, signal drifting, respiratory and cardiac fluctuations, etc., causing a low temporal SNR (tSNR), which could become even lower as the spatial resolution increases. Therefore, proper preprocessing steps are necessary to scrub the raw time series before connectivity analysis. Nevertheless, no consensus has been reached across studies regarding the optimal preprocessing pipeline1. As most sophisticated preprocessing software are tailored for human fMRI2,3, extra difficulties are present on animal studies. In addition, high-resolution rs-fMRI also imposes new requirements and challenges to the correction accuracy. To deal with these challenges, we developed an fMRI preprocessing pipeline featured in thermal noise suppression, spatial blur reduction, and volume- and subject-wise alignment accuracy augmentation. We applied this pipeline to the high-resolution mouse rs-fMRI followed by atlas-based connectivity analysis and obtained high quality resting state networks (RSNs) from large brain regions down to cortical layers.Methods
Animals and scan conditions: Twelve wild type mice (6 male/6 female) were scanned under a protocol approved by the University of Minnesota IACUC. All mice were inducted with 5% isoflurane mixed in O2:N2O (30:70) gas and anesthetized with 1.6% isoflurane during the preparation. After mice were in the magnet, the anesthesia was switched to dexmedetomidine (i.p. 0.3 mg/kg bolus followed by 0.6 mg/kg/hr infusion). Data acquisition started once the respiration rate stabilized at > 140 BPM, and the animals’ physiology was monitored and well controlled throughout the study.MRI experiments and data analysis: The MRI experiments were conducted on a 9.4T/31cm animal scanner (Varian/VNMRJ) using a single loop (1.5cm diameter) surface coil. 2D Gradient echo (GE)-echo planar imaging (EPI) based rs-fMRI data were obtained with TR/TE = 1000/20 ms; matrix size = 96 × 48; FOV = 2.4 cm × 1.2 cm; 12 slices with 0.5 mm thickness. For each mouse, 2-5 rs-fMRI scans with 310 volumes were acquired. T2 weighted anatomical images were acquired with TR/TE=4000/10 ms; matrix = 256 × 128 with the same brain coverage. The rs-fMRI data were preprocessed with the proposed fMRI pipeline shown in Fig. 1. The main features for the functional data processing branch were 1) applying random matrix theory-based principle component analysis (RMT-PCA) denoising with patch processing to effectively remove thermal noise without inducing spatial blur4, 2) minimizing the spatial interpolation steps by combining voxel shifts from various sources such as B1 field distortion, motion, resampling, etc. into one step to avoid further image blurring. The main feature for the subject-level alignment was the flexible and robust registration strategy built on ANTs5 incorporating the averaged undistorted functional images, the anatomical images and (or) brain atlas. Taking advantage of the Allen mouse atlas6, seed-based connectivity analysis was performed on the preprocessed fMRI data to generate RSNs at four hierarchical structure levels.
Results
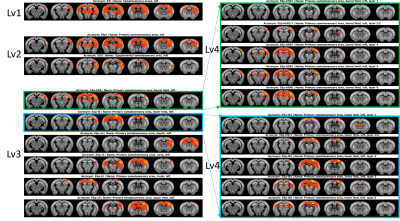
Figure 2 shows the efficacy of RMT-PCA denoising in terms of (A) image quality, (B) temporal signal-to-noise ratio (tSNR) map, and (C) voxel plot7 organized by brain tissues. Dramatic thermal noise was removed from the spatial-temporal data, enhancing the tSNR by about 2.2 times without introducing spatial blur. Figure 3 shows the high quality of non-linearly registered brain atlas to the averaged EPI image of each mouse. Inverse transformation can also be performed to map the EPI images to the original atlas space. Figure 4 shows the default mode networks with seeding at anterior cingulate area (ACA) obtained from (A) original EPI data, and (B) RMT-PCA denoised data. Not only strengthening the overall temporal correlation strength, RMT-PCA denoised data also reveal brain connections that are undetectable with original fMRI data. By seeding somatosensory cortex at various structural levels based on the mouse atlas with the RMT-PCA denoised data (Fig. 5), the higher level RSNs are decomposed into sub-RSNs down to the laminar level with distinguished connection patterns.Discussion and Conclusion
We have developed a robust data preprocessing pipeline with multiple features tailored for high-resolution fMRI brain mapping and demonstrated the high-quality connectomes down to the laminar level in resting mouse brains. The enhanced tSNR brought by the patch-based RMT-PCA denoising promotes functional fidelity, sensitivity, and specificity (Fig. 2 & 4). The optimized preprocessing modules minimize spatial blur and augment the alignment accuracy at volume and subject levels (Fig. 3), in favor of the following group-level functional connectivity analysis. Robust RSNs of mouse brain from large brain areas to thin cortical layers indicate the hierarchical organizations in brain functions (Fig. 5). The distinct patterns in RSNs across cortical layers evidence the non-uniform spontaneous neuronal activities underlay, paving the way to study neural circuits and synchronization. Additionally, the proposed fMRI preprocessing pipeline can also work with high-resolution task fMRI mapping on human and animal brains with minimal modifications, providing a valuable tool to analyze neural circuit and hemodynamic response.Acknowledgements
NIH grants: R01 NS118330, U01 EB026978, R01 MH111413 and P41 EB027061; Keck foundation and Hackett Fund.References
1 Phinyomark, A., Ibanez-Marcelo, E. & Petri, G. Resting-State fMRI Functional Connectivity: Big Data Preprocessing Pipelines and Topological Data Analysis. Ieee T Big Data 2017;3:415-428.
2 Esteban, O. et al. fMRIPrep: a robust preprocessing pipeline for functional MRI. Nat Methods 2019;16:111-116.
3 Glasser, M. F. et al. The minimal preprocessing pipelines for the Human Connectome Project. Neuroimage 2013;80:105-124.
4 Zhu, W. et al. Denoise Functional Magnetic Resonance Imaging with Random Matrix Theory Based Principal Component Analysis. IEEE Trans Biomed Eng 2022;69:3377-3388.
5 Avants, B. B. et al. The Insight ToolKit image registration framework. Frontiers in Neuroinformatics 2014;8.
6 Lein, E. S. et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature 2007;445:168-176.
7 Power, J. D. A simple but useful way to assess fMRI scan qualities. Neuroimage 2017;154:150-158.
Figures
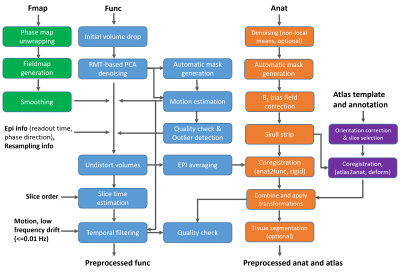
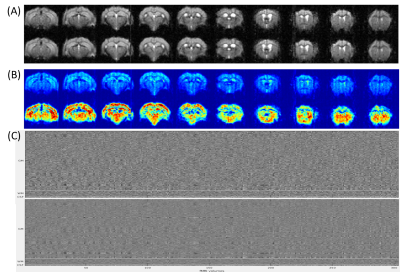
Figure 2. Comparing denoising performance of RMT-PCA in terms of (A) image quality, (B) temporal signal-to-noise ratio (tSNR) map, and (C) voxel plot7. The top row in each panel is the result from raw fMRI data as reference. Dramatic thermal noise was removed from the spatial-temporal data (C), enhancing the tSNR by about 2.2 times (B) without introducing spatial blur (A).
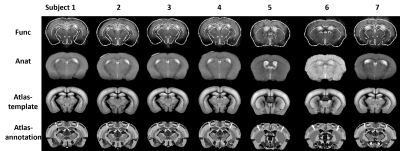
Figure 3. Co-registered Allen mouse brain atlas and anatomical images (Anat) to functional EPI images (Func) for seven randomly selected mice. Rigid transformation was used to register anatomical images to functional images. Non-linear transformation was used to register atlas to anatomical images. Inverse transformation can also be performed to map EPI images to the original atlas space, i.e., subject normalization.
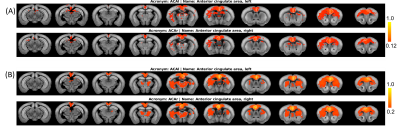
Figure 4. An example of resting state networks with seeding at anterior cingulate area (ACA) obtained from (A) original EPI data, and (B) RMT-PCA denoised data. The default mode networks are observed in both cases, but RMT-PCA denoised data generally show stronger and more robust connections, revealing brain connections that are undetectable with original fMRI data.