0479
Multivariate Classification of fMRI Time Series with Fused Window Transformers
Hasan Atakan Bedel1,2, Irmak Şıvgın1,2, Onat Dalmaz1,2, Salman Ul Hassan Dar1,2, and Tolga Çukur1,2,3
1Department of Electrical and Electronics Engineering, Bilkent University, Ankara, Turkey, 2National Magnetic Resonance Research Center (UMRAM), Bilkent University, Ankara, Turkey, 3Neuroscience Program, Bilkent University, Ankara, Turkey
1Department of Electrical and Electronics Engineering, Bilkent University, Ankara, Turkey, 2National Magnetic Resonance Research Center (UMRAM), Bilkent University, Ankara, Turkey, 3Neuroscience Program, Bilkent University, Ankara, Turkey
Synopsis
Keywords: Data Analysis, fMRI (resting state)
Functional MRI (fMRI) experiments serve a key role in advancing our understanding of human brain function during normal and disease states. Analysis of high-dimensional fMRI data can significantly benefit from recent deep learning approaches, yet existing methods are insufficiently sensitive to the contextual representations in fMRI data across diverse time scales. Here, we present a novel transformer model for fMRI analysis that effectively captures local and global dependencies in fMRI data. Comprehensive demonstrations are provided that show the superior performance of BolT in gender and disease detection against state-of-the-art learning-based methods.Introduction
Functional MRI (fMRI) is a powerful modality that non-invasively records blood-oxygen-level-dependent (BOLD) activity at high spatio-temporal resolution1,8. As such, it is an indispensable tool for neuroscience studies that aim to identify links between multi-variate activity and brain states by analyzing fMRI data recorded during cognitive tasks or resting state.4,7,9. Successful identification relies on the sensitivity of the analysis to contextual dependencies in brain activity across diverse time scales4,6. Given their potential in assessing multi-variate signals, deep learning methods have received recent interest in fMRI analysis2,5,10-17. A first group of methods takes connectivity features as input that are extracted from co-activation patterns of brain regions, and analyzes these features with graph or convolutional networks13,14,15. Although this approach lowers model complexity, connectivity features primarily reflect first-order interactions, disregarding higher-order interactions in brain activity3,4. Another group of methods directly takes brain activity as input, and assesses the information in fMRI data via recurrent or vanilla transformer networks10,11,12. While these methods can improve sensitivity to higher-order interactions, they do not possess explicit mechanisms to simultaneously capture local and global context. Here we introduce a novel transformer model, BolT, for fMRI analysis that directly operates on brain activity for enhanced sensitivity, and leverages a novel fused window attention (FW-MSA) mechanism to capture a hierarchy of local-to-global representations.Methods
BolT: The proposed model leverages a cascade of transformer encoders based on a novel FW-MSA, augmented with token fusion and cross-window regularization (CWR) for integrating representations coherently across fMRI time series (Fig. 1). BolT takes BOLD activity as input and linearly projects the activity at each time instant to obtain a sequence of BOLD tokens and learnable classification tokens (CLS tokens). The output tokens hierarchically encoded by the cascade capture a high-level representation of fMRI data, which is then used in downstream tasks. Individual model elements are described below.a) FW-MSA: Input BOLD tokens, $$$b \in \mathcal{R}^{T \times N}$$$ ($$$T$$$: number of time frames, $$$N$$$: encoding dimensionality), are first split into $$$F=(T-W)/s + 1$$$ overlapping windows of size $$$W$$$ and stride $$$s$$$. Self-attention among base tokens in a given window, and cross-attention between base and fringe tokens from neighboring windows are computed (Fig. 2). A window-specific CLS token is also included in attention calculations. Assuming $$$f_q$$$, $$$f_k$$$ and $$$f_v$$$ are learnable projections for query, key, and value, FW-MSA computations for the $$$i$$$-th window are:
\begin{eqnarray} &&Q_i = f_q( \{ CLS_i, b^{(i \times s)}, ..., b^{(i \times s + W - 1)} \} ) ,\nonumber\\ &&K_i = f_k( \{ CLS_i, b^{(i \times s-L)}, ..., b^{(i \times s + W + L - 1)} \} ), \nonumber\\ &&V_i = f_v( \{ CLS_i, b^{(i \times s-L)}, ..., b^{(i \times s + W + L - 1)} \} ), \nonumber\\ &&\mathrm{Attention}(Q_i, K_i, V_i) = \mathrm{Softmax}(\frac{Q_i K_i^T}{\sqrt{d}} + B) V_i\end{eqnarray}
where $$${B \in \mathbb{R}^{(1+W) \times (1+W+2L)}}$$$ is a learnable position bias17 and $$$d$$$ is the feature dimensionality of the attention head. Note that FW-MSA has linear complexity with sequence length compared to vanilla transformers of quadratic complexity.
b) Token Fusion: The FW-MSA module computes latent representations in each window separately, yet overlapping windows contain common BOLD tokens. Thus, a token fuser is employed to average latent representations of BOLD tokens shared across windows, and to maintain a fixed number of tokens across transformer encoders.
c) CWR: To prevent incoherence among CLS tokens that can degrade classification performance, here high-level representations are regularized across windows in the time series:
\begin{gather} \label{windowConsistencyLoss} L_{CWR} = \frac{1}{N F} \mbox{ } \sum_{i=0}^{F-1} || {CLS}_{i}[M-1] - \frac{1}{F} (\sum_{j=0}^{F-1} {CLS}_{j}[M-1]) ||_2^2,\end{gather}
where $$${CLS_{i}[M-1]}$$$ is the encoded $$${CLS}$$$ token for the $$$i$$$-th window at the output of the last transformer block ($$$M$$$-th).
Datasets: Gender detection in HCP S120018 and disease detection in ABIDE I19 datasets were considered. Resting-state fMRI data from 1093 subjects in HCP S1200, and from 871 subjects in ABIDE I were analyzed. Region extraction was performed using two different brain atlases, AAL21, and Schaefer20. A 10-fold cross-validation procedure was used with a (training, validation, test) split of (80,10,10)%. Model performance was reported on the test set.
Modeling Procedures: Training was performed via the Adam optimizer for 20 epochs, batch size of 32, and initial learning rate of $$$10^{-4}$$$ gradually decreased to $$$10^{-5}$$$. A cross-entropy loss with CWR was used for classification tasks, $$$L_{BolT} = L_{CE} + \lambda L_{CWR}$$$ with $$$\lambda = 0.1$$$.
Results
We demonstrated BolT for gender detection in HCP S1200 and disease detection in ABIDE I, via comparisons against state-of-the-art learning-based and traditional models (Fig. 3). BolT achieves the highest performance with an average improvement in (accuracy, recall, precision, AUC) of (11.80, 14.32, 11.94, 10.17) for gender detection and (5.45, 3.34, 4.75, 5.90) for disease detection over competing methods. Critical brain regions that most significantly contribute to model decisions were derived based on gradient-weighted attention maps17. Critical regions in Fig. 4 for gender detection and Fig. 5 for disease detection closely match recent reports on these brain states16,22,23,24,25.Conclusion
In this study, we introduced a novel transformer model, BolT, for efficient and performant analysis of rich information in fMRI data. Our results indicate that BolT outperforms state-of-the-art methods in gender and disease detection from resting-state fMRI scans. Therefore, BolT holds great promise for sensitive analysis of fMRI time series.Acknowledgements
This study was supported in part by TUBITAK BIDEB, TUBA GEBIP 2015, BAGEP 2017 fellowships, and a TUBITAK 121N029 grant.References
- Hillman, E.M., 2014. Coupling mechanism and significance of the BOLD signal: a status report. Annual Review of Neuroscience 37, 161–181.
- Han, K., Wen, H., Shi, J., Lu, K. H., Zhang, Y., Fu, D., & Liu, Z. (2019). Variational autoencoder: An unsupervised model for encoding and decoding fMRI activity in visual cortex. NeuroImage, 198, 125-136.
- Liu, Z., Rios, C., Zhang, N., Yang, L., Chen, W., & He, B. (2010). Linear and nonlinear relationships between visual stimuli, EEG and BOLD fMRI signals. Neuroimage, 50(3), 1054-1066.
- Deshpande, G., Santhanam, P., & Hu, X. (2011). Instantaneous and causal connectivity in resting state brain networks derived from functional MRI data. Neuroimage, 54(2), 1043-1052.
- Yan, D., Wu, S., Sami, M. T., Almudaifer, A., Jiang, Z., Chen, H., ... & Ma, Y. (2021, December). Improving Brain Dysfunction Prediction by GAN: A Functional-Connectivity Generator Approach. In 2021 IEEE International Conference on Big Data (Big Data) (pp. 1514-1522). IEEE.
- Lowe, M. J., Dzemidzic, M., Lurito, J. T., Mathews, V. P., & Phillips, M. D. (2000). Correlations in low-frequency BOLD fluctuations reflect cortico-cortical connections. Neuroimage, 12(5), 582-587.
- Smith, S. M., Fox, P. T., Miller, K. L., Glahn, D. C., Fox, P. M., Mackay, C. E., ... & Beckmann, C. F. (2009). Correspondence of the brain's functional architecture during activation and rest. Proceedings of the national academy of sciences, 106(31), 13040-13045.
- Feinberg, D. A., Moeller, S., Smith, S. M., Auerbach, E., Ramanna, S., Glasser, M. F., ... & Yacoub, E. (2010). Multiplexed echo planar imaging for sub-second whole brain FMRI and fast diffusion imaging. PloS one, 5(12), e15710.
- Lee, J., Shahram, M., Schwartzman, A., & Pauly, J. M. (2007). Complex data analysis in high‐resolution SSFP fMRI. Magnetic Resonance in Medicine: An Official Journal of the International Society for Magnetic Resonance in Medicine, 57(5), 905-917.
- Malkiel, I., Rosenman, G., Wolf, L., & Hendler, T. (2021). Pre-training and fine-tuning transformers for fmri prediction tasks. arXiv preprint arXiv:2112.05761.
- Nguyen, S., Ng, B., Kaplan, A. D., & Ray, P. (2020, November). Attend and decode: 4d fmri task state decoding using attention models. In Machine Learning for Health (pp. 267-279). PMLR.
- Dvornek, N. C., Ventola, P., Pelphrey, K. A., & Duncan, J. S. (2017, September). Identifying autism from resting-state fMRI using long short-term memory networks. In International Workshop on Machine Learning in Medical Imaging (pp. 362-370). Springer, Cham.
- Kim, B. H., Ye, J. C., & Kim, J. J. (2021). Learning dynamic graph representation of brain connectome with spatio-temporal attention. Advances in Neural Information Processing Systems, 34, 4314-4327.
- Li, X., Zhou, Y., Dvornek, N., Zhang, M., Gao, S., Zhuang, J., Scheinost, D., Staib, L.H., Ventola, P., Duncan, J.S., 2021. Braingnn: Interpretable brain graph neural network for fMRI analysis. Medical Image Analysis 74, 102233.
- Kawahara, J., Brown, C.J., Miller, S.P., Booth, B.G., Chau, V., Grunau, R.E., Zwicker, J.G., Hamarneh, G., 2017. BrainNetCNN: Convolutional neural networks for brain networks; towards predicting neurodevelopment. NeuroImage 146, 1038–1049.
- Abraham, A., Milham, M.P., Di Martino, A., Craddock, R.C., Samaras, D., Thirion, B., Varoquaux, G., 2017. Deriving reproducible biomarkers from multi-site resting-state data: An Autism-based example. NeuroImage 147, 736–745.
- Bedel, H. A., Şıvgın, I., Dalmaz, O., Dar, S. U. H., & Çukur, T. (2022). BolT: Fused Window Transformers for fMRI Time Series Analysis. arXiv preprint arXiv:2205.11578.
- Van Essen, D.C., Smith, S.M., Barch, D.M., Behrens, T.E., Yacoub, E., Ugurbil, K., Consortium, W.M.H., et al., 2013. The WU-Minn human connectome project: an overview. NeuroImage 80, 62–79.
- Di Martino, A., Yan, C.G., Li, Q., Denio, E., Castellanos, F.X., Alaerts, K., Anderson, J.S., Assaf, M., Bookheimer, S.Y., Dapretto, M., et al., 2014. The autism brain imaging data exchange: towards a large-scale evaluation of the intrinsic brain architecture in autism. Molecular psychiatry 19, 659–667.
- Schaefer, A., Kong, R., Gordon, E.M., Laumann, T.O., Zuo, X.N., Holmes, A.J., Eickhoff, S.B., Yeo, B.T., 2018. Local-global parcellation of the human cerebral cortex from intrinsic functional connectivity MRI. Cerebral Cortex 28, 3095–3114.
- Tzourio-Mazoyer, N., Landeau, B., Papathanassiou, D., Crivello, F., Etard, O., Delcroix, N., Mazoyer, B., Joliot, M., 2002. Automated anatomical labeling of activations in SPM using a macroscopic anatomical parcellation of the MNI MRI single-subject brain. NeuroImage 15, 273–289.
- Ritchie, S.J., Cox, S.R., Shen, X., Lombardo, M.V., Reus, L.M., Alloza, C., Harris, M.A., Alderson, H.L., Hunter, S., Neilson, E., et al., 2018. Sex differences in the adult human brain: evidence from 5216 UK biobank participants. Cerebral Cortex 28, 2959–2975
- Filippi, M., Valsasina, P., Misci, P., Falini, A., Comi, G., Rocca, M.A., 2013. The organization of intrinsic brain activity differs between genders: A restingstate fMRI study in a large cohort of young healthy subjects. Human Brain Mapping 34, 1330–1343
- Buckner, R.L., Andrews-Hanna, J.R., Schacter, D.L., 2008. The brain’s default
network: anatomy, function, and relevance to disease. Annals of the New
York Academy of Sciences 1124, 1–38
- Chen, Y.Y., Uljarevic, M., Neal, J., Greening, S., Yim, H., Lee, T.H., 2021. Excessive functional coupling with less variability between salience and default-mode networks in Autism Spectrum Disorder. Biological Psychiatry: Cognitive Neuroscience and Neuroimaging
Figures
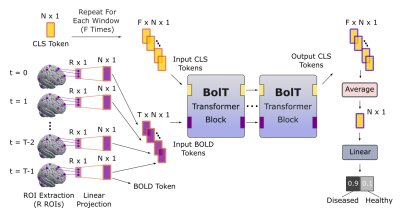
Figure 1: Overview of the proposed BolT model. Four-dimensional fMRI data are subjected to region-of-interest (ROI) extraction based on an anatomical atlas, and then linearly projected to compute input BOLD tokens. Next, a series of transformer blocks processes BOLD tokens across a collection of F temporally-overlapping windows within the time series. The transformer blocks compute latent representations of BOLD and high-level CLS (i.e., classification) tokens; and the CLS tokens are used for the classification task at the output layer.
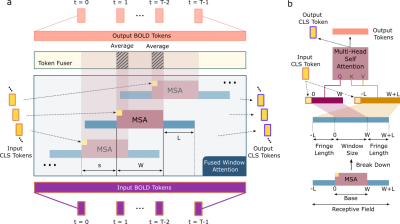
Figure 2: (a) Schematic of the FW-MSA and token fusion modules in the transformer blocks. BOLD tokens are split into a set of overlapping time windows of size W and stride s, along with a window-specific CLS token. Following FW-MSA, attention-derived latent representations are fused across windows for each BOLD token that appears in multiple time windows. (b) Each window has a bilateral fringe region of size L that enables interactions with neighboring windows in FW-MSA.
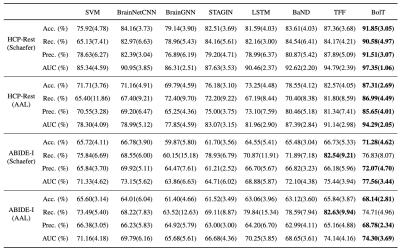
Figure 3: Performance of competing methods for gender detection in HCP S1200 and for disease detection in ABIDE-I. Results are shown for ROI definitions based on Schaefer or AAL atlases. Accuracy, recall, precision and AUC are reported as mean(std) across test folds. The top-performing model is indicated in boldface for each classification task.
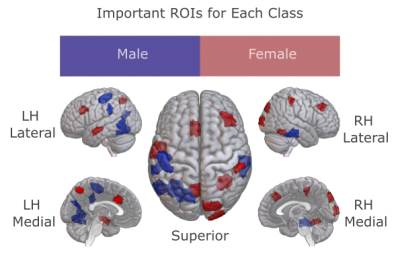
Figure 4: The most influential ROIs for gender detection were visualized for male and female subjects in HCP S1200. Elevated response in a red ROI implies that the individual is female, while elevated response in a blue ROI implies that the individual is male.
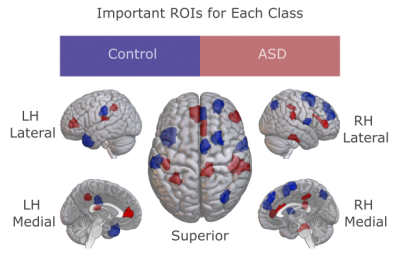
Figure 5: The most influential ROIs for disease detection were visualized for ASD patients and healthy controls in ABIDE-I. Elevated response in a red ROI implies that the individual has ASD, while elevated response in a blue ROI implies that the individual is healthy.
DOI: https://doi.org/10.58530/2023/0479