0437
Using deep learning to identify LNM and LVSI of endometrial cancer from conventional MRI: a preliminary two-center study1Shanghai Key Laboratory of Magnetic Resonance, East China Normal University, Shanghai, China, 2Department of Radiology, Obstetrics and Gynecology Hospital, Fudan University, Shanghai, China, 3Philips Healthcare, Shanghai, China
Synopsis
Keywords: Uterus, Cancer
We developed a multi-task deep learning model using multi-parametric MRI to simultaneously predict lymphatic nodes metastasis (LNM) and lymphatic vascular space invasion (LVSI) in patients with endometrial cancer. Cross-modality attention mechanism was integrated with the model to learn the within and cross modality-specific features which could enhance the performance of network. In this study, we also treated endometrial cancer regions as the anatomical prior knowledge to capture the discriminative information from the whole MR images. The results showed the proposed model predicted LNM and LVSI with a high accuracy in both internal and external test datasets.INTRODUCTION
Endometrial cancer (EC) is the most common gynecological cancer worldwide1 and for lesions with lymphatic nodes metastasis (LNM) and lymphatic vascular space invasion (LVSI), the prognosis is relatively poor. Therefore, identifying LNM and LVSI early and accurately is crucial to treatments planning. MRI is the modality of choice in gynecological cancer staging, due to its high soft-tissue contrast. Thus, we proposed a novel multi-task deep learning model for the simultaneous prediction of LNM and LVSI of endometrial cancer using multi-parametric MRI (mpMRI) images.METHODS
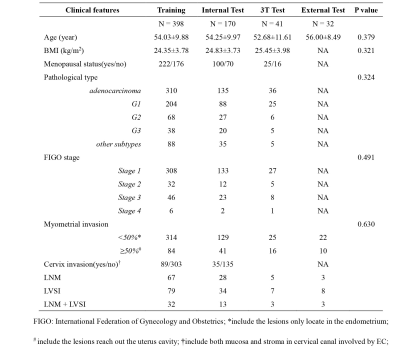
We retrospectively collected 568 EC patients who underwent preoperative MRI on a 1.5T scanner (Magnetom Avanto, Siemens) from January 2015 to April 2019 and randomly split them into a training cohort (N=398) and an internal test cohort (N=170). Forty-one patients (from May 2019 to April 2020) scanned on a 3T scanner (Ingenia 3.0T cx, Philips Medical System, Amsterdam, the Netherlands) in institution 1 and 32 patients scanned on a 1.5T scanner (Optima MR 360, GE) in institution 2 were used as two external test cohorts. The clinical and characteristics of the patients in different cohorts were summarized in Table 1.T2WI, contrast-enhanced T1WI (CE-T1WI) and DWI images were enrolled in this study. Volume of lesion was manually outlined on MRI by an experienced radiologist using ITK-SNAP software. Contrast limited adaptive histogram equalization (CLAHE)2 was used for image enhancement and harmonization. Online random augmentation strategy including shifting, rotation, shearing, vertical flipping, horizontal flipping, elastic and gamma transform was used in training to increase the robustness of model and the sample was resized to 320×320×64 before being fed into the model.
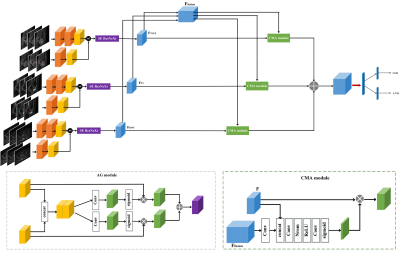
Based on the prior knowledge on EC and mpMRI, we designed a novel multi-task DL network (Figure 1) for the simultaneous prediction of LNM and LVSI in EC patients. Three-dimension ResNeXt3 integrated with squeeze-and-excitation (SE) block was used as the backbone to extract high-level semantic representation from each MRI modality. Anatomical gate (AG)4 was used to help the network to focus on the region around the EC lesion. A cross-modality attention (CMA) module5 was incorporated to fuse features from different MRI modalities to get cross-modality features, which was fused with modality-specific features to predict LNM and LVSI probabilities.
Four-fold cross-validation was used for training. A weighted sum of focal loss6 for LNM (LLNM) and LVSI ( LLVSI) classification was used as loss function. An uncertainty-based method7 was employed to adaptively adjust weights of two losses. The joint loss was described as:
$$\style{font-family:'Times New Roman'}{L_{joint}=\frac1{2{\sigma^2}_{LNM}}L_{LNM}+\frac1{2{\sigma^2}_{LVSI}}L_{LVSI}+log\sigma_{LNM}\sigma_{LVSI}}$$
where σLNM and σLVSI were learnable weights updated adaptively during training. Adam optimizer with ReduceLRonPlateau scheduler was used as optimizer. The batch size was set to 4 and if the validation loss did not decrease over 20 epochs, the training was early-stopped. The proposed network was implemented with PyTorch 1.9.0 and trained on a workstation equipped with three NVIDIA A100 GPUs with 40 GB memory each.
RESULTS
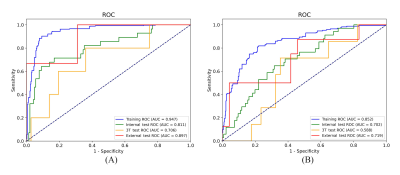
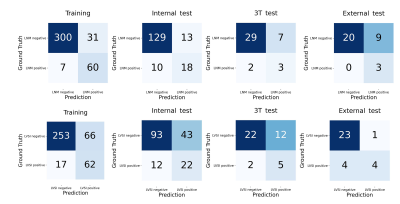
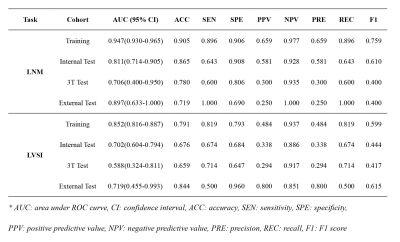
Figure 2 shows the ROC curves for the prediction of LNM and LVSI in different cohorts and we also used confusion matrix to evaluate the performance of the proposed model in Figure 3. For LNM prediction, the model achieved AUC values of 0.947 (95% CI, 0.930-0.965), 0.811 (95% CI, 0.714-0.905), 0.706 (95% CI, 0.400-0.950) and 0.897 (95% CI, 0.633-1.000) in the training, internal test, 3T test and external test cohorts, respectively. For LVSI prediction, the model yielded AUC values of 0.852 (95% CI, 0.816-0.887), 0.702 (95% CI, 0.604-0.794), 0.588 (95% CI, 0.324-0.811) and 0.719 (95% CI, 0.455-0.993) in the training, internal test, 3T test and external test cohorts, respectively. Detailed metrics of predictive performance of the proposed model are listed in Table 2.DISCUSSION AND CONCLUSION
In this study, we proposed a novel network to simultaneously identify LNM and LVSI in EC patients. Radiomics has been used to study LNM of EC8-10, however, we found no study on AI modeling for LVSI of EC. For LNM identification, our trained model achieved a test AUC of 0.811, higher than those of radiomics models reported by previous studies, which were in the range of 0.730 to 0.7628-10. The better performance of the proposed model can be attributed to the larger dataset we used to train the model, and to the fact that the proposed model simultaneously handles two different clinical tasks, which gives the model more constraints to learn both task-specific and shared features, making it less likely to overfit.Furthermore, we also made full use of the prior knowledge on mpMRI-based EC diagnosis in the design of network architecture: 1. CMA module was used to adaptively aggregate the modality-specific features to learn the discriminative cross-modality information in mpMRI; 2. Anatomical gating was used to help the model focus on the lesion. Besides, a joint loss function with learnable weights was used to balance the performance of LNM and LVSI identification tasks in the training process.
In summary, our proposed DL model achieved high performance in both LNM and LVSI identification based on mpMRI and it has the potential to help clinicians for better diagnosis and treatment planning for EC patients.
Acknowledgements
This project is supported by National Natural Science Foundation of China (61731009, 81771816) and the Open Project of Shanghai Key Laboratory of Magnetic Resonance.References
1. Keles DK, Evrimler S, Merd N, Erdemoglu E. Endometrial cancer: the role of MRI quantitative assessment in preoperative staging and risk stratification. Acta Radiol. 2022;63,1126-1133.
2. Reza, A.M. Realization of the Contrast Limited Adaptive Histogram Equalization (CLAHE) for Real-Time Image Enhancement. The Journal of VLSI Signal Processing-Systems for Signal, Image, and Video Technology. 2004;38,35–44.
3. Saining X, Ross Girshick, Piotr Dollar, et al. Aggregated Residual Transformations for Deep Neural Networks. Proceedings of the IEEE conference on computer vision and pattern recognition. 2017;1492-1500.
4. Sun L, Shao W, Zhang D, et al. Anatomical Attention Guided Deep Networks for ROI Segmentation of Brain MR Images. IEEE Transactions on Medical Imaging.2019;99,1-1.
5. Yao Zhang, Jiawei Yang, Jiang Tian, et al. Modality-Aware Mutual Learning for Multi-modal Medical Image Segmentation. Medical Image Computing and Computer Assisted Intervention-MCCAI.2021;12901.
6. Lin T Y, Goyal P, Girshick R, et al. Focal Loss for Dense Object Detection. 2017 IEEE International Conference on Computer Vision (ICCV). 2017; 2999-3007.
7. R. Cipolla, Y. Gal, and A. Kendall. Multi-task learning using uncertainty to weigh losses for scene geometry and semantics, 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition.2018; 7482–7491.
8. Qu, J., Shen, C., Qin, J. et al. The MR radiomic signature can predict preoperative lymph node metastasis in patients with esophageal cancer. Eur Radiol. 2019;29,906–914.
9. Ytre-Hauge S, Dybvik JA, Lundervold A, et al. Preoperative tumor texture analysis on MRI predicts high-risk disease and reduced survival in endometrial cancer. J Magn Reson Imaging. 2018;48(6),1637-1647.
10. Xu X, Li H, Wang S, et al. Multiplanar MRI-Based Predictive Model for Preoperative Assessment of Lymph Node Metastasis in Endometrial Cancer. Front Oncol. 2019;9,1007.
Figures