0430
Synthesizing Complex Multicoil MRI Data from Magnitude-only Images1Radiology and Biomedical Imaging, University of California, San Francisco, San Francisco, CA, United States, 2UC Berkeley - UCSF Graduate Program in Bioengineering, Berkeley and San Francisco, CA, United States, 3Electrical Engineering and Computer Sciences, University of California, Berkeley, Berkeley, CA, United States
Synopsis
Keywords: Machine Learning/Artificial Intelligence, Machine Learning/Artificial Intelligence
Obtaining paired, diverse and expert annotated medical data is extremely challenging especially for MRI reconstruction since raw data, including phase information is typically discarded from the scan leaving only the magnitude image for clinical assessment and diagnosis. Phase information contains valuable information about physiology, pathology and other tissue characteristics that are useful in developing more robust deep learning MRI reconstruction methods. In this work, we show that state of the art physics-based image reconstruction networks trained on synthetic raw MRI data consisting of synthetic phase and coil information perform comparably to image reconstruction networks trained on ground truth k-space data.Introduction
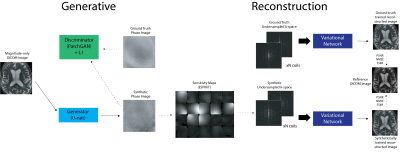
Despite the proliferation of deep learning for accelerated acquisition and enhanced image reconstruction, aggregation of large and diverse MRI datasets continues to pose a barrier for effective clinical translation of these technologies. One major challenge is in retaining the MRI phase (required for image reconstruction) in clinical scanning, as only magnitude images are typically saved and used for clinical assessment and diagnosis. While academic medical centers have begun the long concerted effort to upgrade clinical database systems and overcome regulatory and standardization issues, current databases have limitations including lack of diversity in anatomy and pathology, lack of diversity of acquisition types, and contained preprocessed magnitude-only data which can lead to biased results1 all of which limit the generalization potential and clinical viability of advanced reconstruction models.2To address this, we present a method for synthesizing realistic MRI that is complex-valued and multi-channel from magnitude-only images, allowing tasks involving MRI reconstruction to leverage large datasets contained in DICOM objects that are typically magnitude-only. Our method, based on the pix2pix3 framework, generates synthetic phase images from input magnitude images and ESPIRiT4 sensitivity maps derived from prior data to generate multi-channel data. It was evaluated by training a Variational Network5 trained on ground truth data and synthetic data, undersampled in k-space. We show that state of the art physics-based image reconstruction networks trained on synthetic raw MRI data consisting of synthetic phase and coil information perform comparably to image reconstruction networks trained on ground truth k-space data.
Methods
We used 16-coil (22691 training, 6541 test) and 20-coil data (18519 training, 5485 test) from the fastMRI dataset6 consisting of raw k-space with corresponding T1-weighted, T2-weighted, and FLAIR contrast brain images acquired at 1.5T and 3T with a fast spin echo (FSE) pulse sequence with an echo train length (ETL) of 4. To generate realistic synthetic phase images from magnitude-only input images, we trained a conditional GAN consisting of a 16-layer U-Net generator with an encoder-decoder architecture with skip connections. The PatchGAN3 discriminator decides if a 70x70 patch in the generated image is real or fake. The model uses a hybrid objective consisting of two loss functions: an adversarial loss and a regularized l1 distance loss function (to encourage less blurring).\begin{equation} \label{pix2pixobj} G^{*} = arg\min_{G}\max_{D} \underbrace{\mathbb{E}_{x,y}[logD(x,y)] + \mathbb{E}_{x,z}[log(1 - D(x, G(x,z))]}_{\mathcal{L}_{cGAN}} + \lambda \underbrace{\mathbb{E}_{x,y,z}[ \|y-G(x,z) \|_{1}]}_{\mathcal{L}_{L1}}\end{equation}
To evaluate the utility of synthetic multicoil k-space for image reconstruction, multiple equispaced undersampling masks (4x, 8x, 16, 20x acceleration factors) with a center fraction of 0.04 were applied to k-space. Two Variational Networks5 were then trained for each 16-coil and 20-coil dataset, each on synthetic and ground truth multicoil k-space. Each trained image reconstruction model was then run on the same ground-truth test set (235 for 16-coil dataset and 214 for 20-coil dataset) and the quality of reconstructed magnitude images was evaluated using standard quantitative image reconstruction metrics: PSNR, SSIM, NMSE.
Results and Discussion
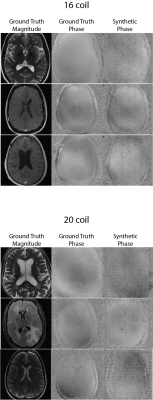
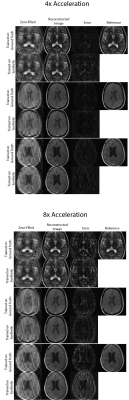
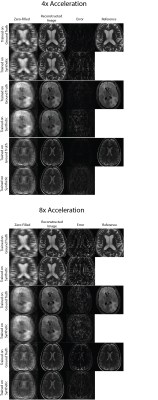
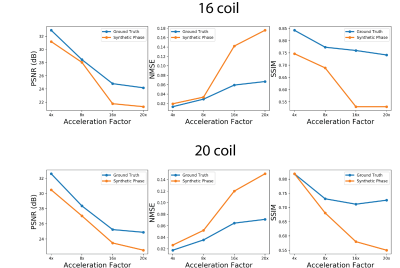
Figures 2 shows sample comparisons between synthetic and ground truth phase images and Figures 3 and 4 show images reconstructed with Variational Networks models trained with undersampled ground truth and synthetic k-space. The synthetic phase images show several expected features, including low spatial-frequency components and some tissue contrast seen between the ventricles. In some cases, there are blocking artifacts possibly resulting from the PatchGAN discriminator. At 4x acceleration, the error maps of the reconstructed images trained on synthetic data are slightly higher compared to the images trained on a ground-truth model, although visually there are no obvious artifacts in either method. Additionally we can see more errors in high resolution features, possibly due to the lack of high frequency details in the synthetic phase images. These lack of details could also manifest during image reconstruction where the model tries to "fill in" features that it never sees.Figure 5 compares Variational Network reconstruction performance on models trained on ground-truth and synthetic data at different acceleration factors. From the plots, variational networks trained on undersampled synthetic data perform comparably to the same model trained on ground truth undersampled k-space at 4x as measured by PSNR, NMSE and SSIM. At 8x acceleration factor, the synthetically trained models' performance dips but remains relatively comparable to the ground-truth trained models performance. At 16x and 20x acceleration factors however, synthetically trained models' performance is worse in all three categories.
Conclusion
This work presents a new method for synthesizing realistic MRI data that is complex-valued and multi-channel from magnitude-only images, which were evaluated by comparing the reconstruction performance of Variational Networks trained on ground-truth k-space and synthetic k-space. Our results suggest that this approach for generating synthetic phase and multicoil data is adequate for training MRI reconstruction models at lower acceleration factors (4x, 8x), shown by the Variational Networks results.We believe a major opportunity for such a synthetic data pipeline is to generate multicoil k-space from clinical magnitude-only scans where the raw data has been discarded and use this synthetic data to train multi-task reconstruction networks6 to take advantage of existing clinical annotations for downstream tasks, enabling creation of customized datasets for machine learning that includes image reconstruction tasks.
Acknowledgements
No acknowledgement found.References
1. Efrat Shimron, Jonathan I. Tamir, Ke Wang, and Michael Lustig. Implicit data crimes: Machine learning bias arising from misuse of public data. Proceedings of the National Academy of Sciences, 119(13), March 2022.
2. Kerstin Hammernik, Thomas Ku ̈stner, Burhaneddin Yaman, Zhengnan Huang, Daniel Rueckert, Florian Knoll, and Mehmet Akc ̧akaya. Physics-driven deep learning for computational magnetic resonance imaging, 2022.
3. Phillip Isola, Jun-Yan Zhu, Tinghui Zhou, and Alexei A. Efros. Image-to-image translation with conditional adversarial networks. In 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR). IEEE, July 2017.
4. Martin Uecker, Peng Lai, Mark J. Murphy, Patrick Virtue, Michael Elad, John M. Pauly, Shreyas S. Vasanawala, and Michael Lustig. ESPIRiT-an eigenvalue approach to autocalibrating parallel MRI: Where SENSE meets GRAPPA. Magnetic Resonance in Medicine, 71(3):990–1001, May 2013.
5.Kerstin Hammernik, Teresa Klatzer, Erich Kobler, Michael P. Recht, Daniel K. Sodickson, Thomas Pock, and Florian Knoll. Learning a variational network for reconstruction of accelerated MRI data. Magnetic Resonance in Medicine, 79(6):3055–3071, November 2017.
6.Florian Knoll, Tullie Murrell, Anuroop Sriram, Nafissa Yakubova, Jure Zbontar, Michael Rabbat, Aaron De- fazio, Matthew J. Muckley, Daniel K. Sodickson, C. Lawrence Zitnick, and Michael P. Recht. Advancing machine learning for MR image reconstruction with an open competition: Overview of the 2019 fastMRI challenge. Magnetic Resonance in Medicine, 84(6):3054–3070, June 2020.
7. Francesco Caliva’, Rutwik Shah, Upasana Upadhyay Bharadwaj, Sharmila Majumdar, Peder Larson, and Valentina Pedoia. Breaking speed limits with simultaneous ultra-fast {mri} reconstruction and tissue seg- mentation. In Medical Imaging with Deep Learning, 2020.
Figures