0421
MR Fingerprinting with a Deep Image Prior Reconstruction for Combined T1, T2, and M0 Mapping and Multi-Contrast Cine Imaging1Radiology, University of Michigan, Ann Arbor, MI, United States, 2Biomedical Engineering, University of Michigan, Ann Arbor, MI, United States, 3Harrington Heart and Vascular Institute, University Hospitals Cleveland Medical Center, Cleveland, OH, United States, 4Medicine, Case Western Reserve University, Cleveland, OH, United States
Synopsis
Keywords: MR Fingerprinting/Synthetic MR, Cardiovascular, Quantitative Imaging
This work introduces a self-supervised deep learning reconstruction for cine Magnetic Resonance Fingerprinting, allowing for simultaneous cardiac phase-resolved T1, T2, and M0 mapping (without motion correction or averaging of data across different phases) and bright-blood and dark-blood cine imaging during a 10-second breathhold, with a temporal resolution (24 phases) comparable to standard cine imaging. Results are presented in simulations using the XCAT phantom and in healthy subjects, where the proposed reconstruction yielded reduced noise, undersampling artifacts, and motion blurring compared to previous low-rank and motion-corrected methods.Introduction
Previous work with cine Magnetic Resonance Fingerprinting (MRF) (1,2) employed nonrigid cardiac motion correction that may be prone to errors since images have low SNR, residual aliasing, and variable contrasts. Motion correction may also preclude measurement of dynamic T1-T2 changes. This study proposes an improved cine MRF reconstruction using a deep image prior for true cardiac phase-resolved (without motion correction/averaging) T1, T2, and M0 mapping and bright/dark-blood cine imaging.Methods
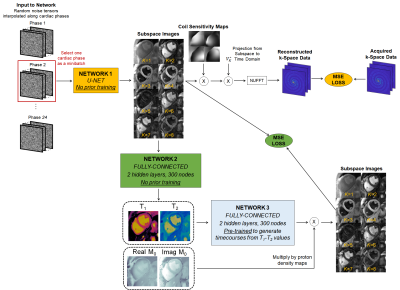
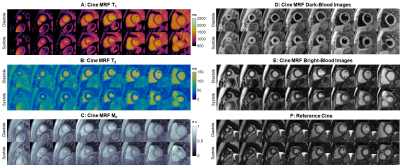
Acquisition/Reconstruction: Data were acquired using a FISP readout with 4-15° flip angles, periodic inversions/T2-preparations, and 1820 TRs during a 10-second breathhold similar to previous work (2). Data were retrospectively gated to 24 cardiac phases using the ECG signal. One dictionary was precomputed for all subjects using a Bloch equation simulation with corrections for slice profile and preparation efficiency (3).This study generalizes a deep image prior (DIP) reconstruction first proposed for diastolic MRF mapping (4,5). Training is performed de novo for each scan, without additional training data (Figure 1). Network 1 (u-net) generates images in a low-dimensional subspace derived from the SVD-compressed dictionary. The input is a random noise tensor (one per cardiac phase); these are randomly initialized for the first and last phases and linearly interpolated for intermediate phases. Network 1 is updated by simulating the forward encoding model and minimizing the MSE loss compared to the acquired density-compensated k-space data. In parallel, subspace images are input to Network 2 (fully-connected) to generate quantitative maps. Network 2 is updated by using the maps to calculate synthetic images that are compared to images generated by Network 1; this step is performed efficiently using a pre-trained “fingerprint generator” (Network 3) instead of a Bloch simulation. Subspace images corresponding to the 2nd and 3rd singular values are used as bright/dark-blood cine images.
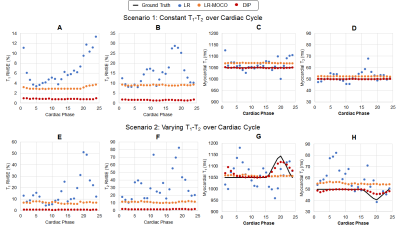
Simulations: Data were simulated using the XCAT phantom (6) with 8-channel sensitivity maps and 60bpm cardiac motion. Two scenarios were tested: (Scenario 1) constant myocardial T1=1050ms and T2=50ms, and (Scenario 2) periodic variations every cardiac cycle with T1 1050-1150ms and T2 40-50ms. Cine MRF maps were reconstructed using three methods: (LR) a low-rank subspace reconstruction with total variation penalties along spatial and cardiac phase dimensions, (LR-MOCO) LR followed by nonrigid motion correction as in (2), and (DIP) the proposed method. RMSE values were computed relative to ground truth maps
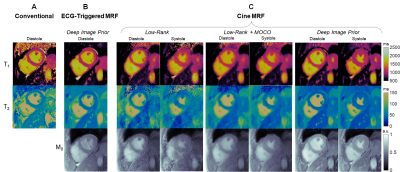
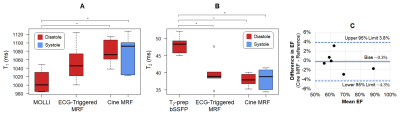
In Vivo: Six healthy subjects were scanned at 1.5T (Sola, MAGNETOM Siemens) at a mid-ventricular slice. Cine MRF data (1.5x1.5x8 mm3, 192x192 matrix) were reconstructed using LR, LR-MOCO, and DIP. Diastolic maps were separately acquired using MRF with prospective triggering and reconstructed using a deep image prior (5). MOLLI (7) and T2-prepared bSSFP (8) maps and reference bSSFP cine images (FA 75°, TR/TE 4.2/2.4ms, 25 phases, 7s breathhold) were collected. T1 and T2 in the left ventricular (LV) septum were compared using Kruskal-Wallis tests with Bonferroni post-hoc corrections, with T1/T2 measured at peak diastole and systole for cine MRF. Single-slice ejection fraction (EF) was assessed using a Bland-Altman test. In one subject, scans were collected over multiple short-axis slices to compare end-diastolic volume (EDV), end-systolic volume (ESV), and EF over the entire LV.
Results
DIP yielded the lowest RMSE in simulations (Figure 2). For Scenario 1 (constant T1-T2), RMSE values averaged over all phases were: LR (6.6% T1, 15.0% T2), LR-MOCO (3.0% T1, 9.0% T2), and DIP (0.9% T1, 1.6% T2). For Scenario 2, DIP detected dynamic T1-T2 changes, although it overestimated the minimum T2 and underestimated the maximum T1. DIP outperformed LR and LR-MOCO, the latter incorrectly estimating constant T1-T2 over all phases.Figure 3 shows quantitative maps using cine MRF with different reconstructions. LR exhibited noise enhancement, while LR-MOCO reduced noise at the expense of motion blurring. DIP yielded the best suppression of noise and undersampling artifacts with minimal blurring. Mean T1 values over all subjects (Figure 4) were: MOLLI (1008ms), ECG-triggered MRF (1052ms), cine MRF with DIP (diastole 1077ms, systole 1076ms). Mean T2: T2-prep bSSFP (48.1ms), ECG-triggered MRF (39.8ms), cine MRF with DIP (diastole 37.9ms, systole 38.2ms). MOLLI T1 was significantly lower than cine MRF. T2-prep bSSFP values were significantly higher than both MRF scans. Single-slice EF using cine MRF with DIP agreed with reference values (mean bias -0.3%; 95% limits of agreement -4.3% to 3.8%). Figure 5 shows a short-axis stack in one subject with EDV 179mL, ESV 48mL, LVEF 73.1% for cine MRF and EDV 198mL, ESV 59mL, LVEF 70.3% for the reference.
Discussion and Conclusions
This study proposed a self-supervised deep learning reconstruction for cine MRF that reduced noise and undersampling artifacts compared to previous low-rank reconstructions. Cardiac phase-resolved maps (without motion correction or registering images over different phases) and multi-contrast cine images were acquired in a single breathhold. While this study used subspace images directly as bright/dark-blood images, synthetic contrasts could be simulated from tissue property maps. This work may enable streamlined exams by assessing function and tissue properties during an all-in-one approach. A current limitation is the 8-hour computation time on a GPU. Future work will include validation in patients expected to have abnormal T1/T2 and cardiac function, comparison to the generalized low-rank reconstruction proposed in (9), and exploration of potential changes in T1-T2 over the cardiac cycle.Acknowledgements
This work was supported by the Michigan Institute for Clinical & Health Research (MICHR) Grant UL1TR002240, Siemens Healthineers, and National Institutes of Health / National Heart, Lung, and Blood Institute (NIH/NHLBI) R01HL163030 and R01HL153034.References
1. Jaubert O, Cruz G, Bustin A, et al. Free-running cardiac magnetic resonance fingerprinting: Joint T1/T2 map and Cine imaging. Magn. Reson. Imaging 2020;68:173–182.
2. Hamilton JI, Jiang Y, Eck B, Griswold M, Seiberlich N. Cardiac cine magnetic resonance fingerprinting for combined ejection fraction, T1 and T2 quantification. NMR Biomed. 2020;33:e4323.
3. Hamilton JI, Jiang Y, Ma D, et al. Investigating and reducing the effects of confounding factors for robust T1 and T2 mapping with cardiac MR fingerprinting. Magn. Reson. Imaging 2018;53:40–51.
4. Ulyanov D, Vedaldi A, Lempitsky V. Deep Image Prior. In: Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition. IEEE Computer Society; 2018. pp. 9446–9454.
5. Hamilton JI. A Self-Supervised Deep Learning Reconstruction for Shortening the Breathhold and Acquisition Window in Cardiac Magnetic Resonance Fingerprinting. Front. Cardiovasc. Med. 2022;9:928546.
6. Segars WP, Sturgeon G, Mendonca S, Grimes J, Tsui BMW. 4D XCAT phantom for multimodality imaging research. Med. Phys. 2010;37:4902–4915.
7. Messroghli DR, Radjenovic A, Kozerke S, Higgins DM, Sivananthan MU, Ridgway JP. Modified Look-Locker inversion recovery (MOLLI) for high-resolution T1 mapping of the heart. Magn. Reson. Med. 2004;52:141–146.
8. Giri S, Chung YC, Merchant A, et al. T2 quantification for improved detection of myocardial edema. J. Cardiovasc. Magn. Reson. 2009;11.
9. Cruz G, Qi H, Jaubert O, et al. Generalized low-rank nonrigid motion-corrected reconstruction for MR fingerprinting. Magn. Reson. Med. 2022;87:746–763.
Figures