0405
Systematic evaluation of the robustness of open-source networks for MRI multicoil reconstruction1Graduate School of Science and Technology, University of Tsukuba, Tsukuba, Japan, 2FUJIFILM Healthcare Corporation, Tokyo, Japan
Synopsis
Keywords: Image Reconstruction, Machine Learning/Artificial Intelligence, Deep Learning Reconstrcution
Deep neural networks (DNNs) for MRI reconstruction often require large datasets for training, but in clinical settings, the domains of datasets are diverse, and the degree to which deep neural networks are the robustness of DNNs to domain differences between training and testing datasets has been an open question. Here, we evaluated the robustness of four open-source multicoil networks to differences in the domain. We found that model-based networks exhibit higher robustness than data-driven networks and that robustness varies across network architectures, even within model-based networks. Our results provide insight into what network architectures are effective for generalization performance.Introduction
Deep learning (DL)-based MRI reconstruction has the potential for high performance [1]–[3]. Supervised learning models are commonly used [4], and the performance is higher when the dataset domain used for testing (target domain) is closer to the dataset domain used for training (source domain). However, clinical MRI datasets can have various domain differences depending on the facility, scanner, anatomy, sequence, etc. It is practically difficult to prepare training datasets that encompass such diversity, and differences in the distribution of source and target domains are inevitable.Thus, there is an open question about how robust the proposed reconstruction networks are against these domain differences. To answer this question, reports [5]–[7] have examined the generalization performance of reconstruction networks. In these studies, however, comparisons have been made only among limited categories of models and domain types.
Here we evaluate the generalization of the reconstruction networks across various domains under clinically practical conditions. Specifically, we compare the reconstruction performance between four network models in terms of the number of images, sampling pattern, acceleration factor (AF), noise level, contrast, and anatomical structure. We use publicly-available networks and multicoil datasets to provide research data that is easy to follow up.
Method
Deep neural networksWe used two categories of MRI reconstruction networks: data-driven and model-based networks. The former uses a large amount of data to learn potential mapping relationships from input (zero-filled image or undersampled k-space data) to output artifact-free images. The latter uses a cascade of DNN to unroll the traditional non-DL optimization iteration algorithm. The models used were as follows:
Data-driven model
- U-Net [8]
Model-based model
- Deep Cascade of Convolutional Neural network (DC-CNN) [3]
- Hybrid Cascade [9]
- Variational Network (VarNet) [1]
Traditional iterative method
CG-SENSE [10], [11]
We used mean-squared error (MSE) for training as the loss function and Adam as the optimizer. For U-net, we used the code from the FastMRI Challenge implementation [12] and trained with 200 epochs, and learning-rate was $$$1.0\times10^{-3}$$$. For DC-CNN and Hybrid Cascade, we modified a public code [13] to use the multicoil data and trained with 50 epochs, and the learning-rate was $$$1.0\times10^{-4}$$$. For VarNet, we also used a public code [14] and trained with 50 epochs, and the learning-rate was $$$1.0\times10^{-3}$$$. For CS-SENSE, we used the reconstruction code of Sigpy [15] with the L1-Wavelet.
Dataset and generalization evaluation
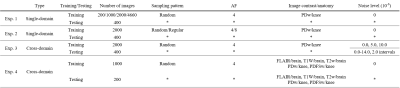
We used the public FastMRI dataset [12]. The multicoil k-space data was retrospectively undersampled and used as input. The sensitivity map was estimated from the undersampled data by ESPiRIT [16], and also used as input. We conducted the following single-domain test (dataset domains for training and testing were the same) and cross-domain test (dataset domains were different between training and testing), Experiments 1-4. The detailed conditions are listed in Table 1.
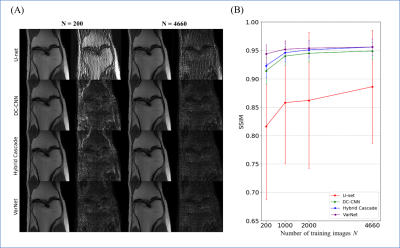
Exp. 1: Number of training image (single-domain test)
The number of training images was changed from 200 to 4660 images, and the learned models were tested for 400 images.
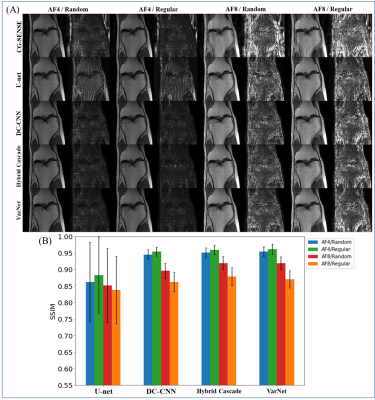
Exp. 2: Sampling pattern and AF (single-domain test)
Four sampling patterns were used. The patterns for training and testing were the same.
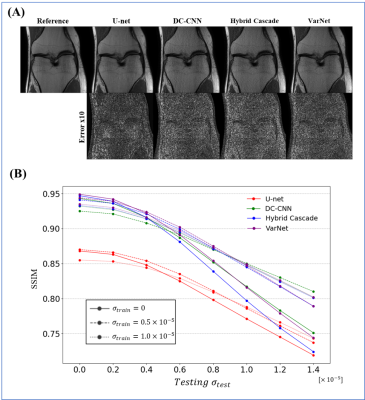
Exp. 3: Noise level (cross-domain test)
Gaussian noises were added to train and test datasets with different noise levels.
Exp. 4: Contrast and anatomy (cross-domain test)
We trained with five subsets with different image contrasts and anatomies. We cross-tested against a different subset than the subset used for training.
Results
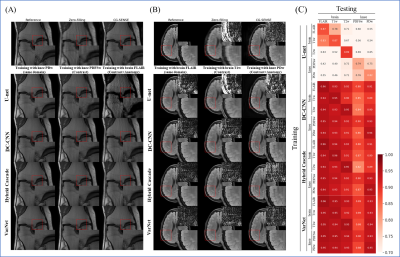
The results for single-domain tests (Exp. 1 and 2) are shown in Figs. 1 and 2. VarNet performed better than other networks regardless of the number of training images. The model-based networks (VarNet, Hybrid Cascade, and DC-CNN) outperformed the data-driven network (U-Net).The results for the cross-domain test (Exp. 3) are shown in Fig. 3. All the model-based networks performed better than U-Net. U-Net was most robust to noise-level differences, although it exhibited worse metrics within the range of noise differences used in this study. Among the model-based networks, Hybrid Cascade was the least robust to noise difference, followed by VarNet.
The results for the cross-domain test (Exp. 4) are shown in Fig. 4. VarNet was robust to both contrast and anatomy domain differences. Hybrid Cascade and DC-CNN were robust only to contrast differences. U-Net was not robust in any case.
Discussion
VarNet generally showed the highest robustness among the model-based networks in all tests. Since VarNet uses an iterative gradient descent algorithm, it could have extracted features independent of differences between training and target domains.Hybrid Cascade showed higher robustness than DC-CNN except for the noise test. The difference between DC-CNN and Hybrid Cascade is that the latter has a cascade CNN in the k-space domain. The result shows that using the k-space CNN effectively improves performance at the cost of potentially reduced robustness to noise levels.
U-Net showed the lowest robustness in all tests. This is probably because U-Net does not have the regularization term and iterative building blocks of the model-based reconstruction models.
Conclusion
Here we validated the generalization performance of multiple open-source networks for MRI reconstruction through single- and cross-domain tests. Our results provide insight into the architectures that work effectively for generalization performance.Acknowledgements
No acknowledgement found.References
[1] K. Hammernik et al., “Learning a variational network for reconstruction of accelerated MRI data,” Magn. Reson. Med., vol. 79, no. 6, pp. 3055–3071, 2018.
[2] H. K. Aggarwal, M. P. Mani, and M. Jacob, “MoDL: Model-based deep learning architecture for inverse problems,” IEEE Trans. Med. Imaging, vol. 38, no. 2, pp. 394–405, 2018.
[3] J. Schlemper, J. Caballero, J. V. Hajnal, A. N. Price, and D. Rueckert, “A deep cascade of convolutional neural networks for dynamic MR image reconstruction,” IEEE Trans. Med. Imaging, vol. 37, no. 2, pp. 491–503, 2017.
[4] Y. Chen et al., “AI-based reconstruction for fast MRI—a systematic review and meta-analysis,” Proc. IEEE, vol. 110, no. 2, pp. 224–245, 2022.
[5] K. Hammernik, J. Schlemper, C. Qin, J. Duan, R. M. Summers, and D. Rueckert, “Systematic evaluation of iterative deep neural networks for fast parallel MRI reconstruction with sensitivity-weighted coil combination,” Magn. Reson. Med., vol. 86, no. 4, pp. 1859–1872, 2021.
[6] J. Huang, S. Wang, G. Zhou, W. Hu, and G. Yu, “Evaluation on the generalization of a learned convolutional neural network for MRI reconstruction,” Magn. Reson. Imaging, vol. 87, pp. 38–46, 2022.
[7] “Assessment of the generalization of learned image reconstruction and the potential for transfer learning - Knoll - 2019 - Magnetic Resonance in Medicine - Wiley Online Library.” https://onlinelibrary.wiley.com/doi/full/10.1002/mrm.27355 (accessed Mar. 15, 2022).
[8] O. Ronneberger, P. Fischer, and T. Brox, “U-Net: Convolutional networks for biomedical image segmentation,” in International Conference on Medical image computing and computer-assisted intervention, 2015, pp. 234–241.
[9] R. Souza, R. M. Lebel, and R. Frayne, “A hybrid, dual domain, cascade of convolutional neural networks for magnetic resonance image reconstruction,” in International Conference on Medical Imaging with Deep Learning, 2019, pp. 437–446.
[10] K. P. Pruessmann, M. Weiger, M. B. Scheidegger, and P. Boesiger, “SENSE: sensitivity encoding for fast MRI,” Magn. Reson. Med. Off. J. Int. Soc. Magn. Reson. Med., vol. 42, no. 5, pp. 952–962, 1999.
[11] K. P. Pruessmann, M. Weiger, P. Börnert, and P. Boesiger, “Advances in sensitivity encoding with arbitrary k-space trajectories,” Magn. Reson. Med. Off. J. Int. Soc. Magn. Reson. Med., vol. 46, no. 4, pp. 638–651, 2001.
[12] J. Zbontar et al., “fastMRI: An open dataset and benchmarks for accelerated MRI,” ArXiv Prepr. ArXiv181108839, 2018.
[13] R. Souza, “CD-Deep-Cascade-MR-Reconstruction.” Aug. 11, 2022. Accessed: Nov. 05, 2022. [Online]. Available: https://github.com/rmsouza01/CD-Deep-Cascade-MR-Reconstruction/blob/1ac44ecefbbe229a3359a6101b482af157a80662/README.md
[14] rixez, “Pytorch implementation of Variational Network for Magnetic Resonance Image (MRI) Reconstruction.” Oct. 02, 2022. Accessed: Nov. 05, 2022. [Online]. Available: https://github.com/rixez/pytorch_mri_variationalnetwork
[15] J. B. Martin, F. Ong, J. Ma, J. I. Tamir, M. Lustig, and W. A. Grissom, “SigPy. RF: comprehensive open-source RF pulse design tools for reproducible research,” in Proc. 28th Annual Meeting of ISMRM, 2020.
[16] M. Uecker et al., “ESPIRiT—an eigenvalue approach to autocalibrating parallel MRI: where SENSE meets GRAPPA,” Magn. Reson. Med., vol. 71, no. 3, pp. 990–1001, 2014.
Figures