0404
M4Raw: A Multi-Contrast Multi-Repetition Multi-Channel Raw K-space Dataset for Low-Field MRI Reconstruction1College of Health Science and Environmental Engineering, Shenzhen Technology University, Shenzhen, China, 2Guangdong-Hongkong-Macau Institute of CNS Regeneration, Key Laboratory of CNS Regeneration (Ministry of Education), Jinan University, Guangzhou, China, 3Laboratory of Biomedical Imaging and Signal Processing, the University of Hong Kong, Hong Kong, China, 4Department of Electrical and Electronic Engineering, the University of Hong Kong, Hong Kong, China
Synopsis
Keywords: Machine Learning/Artificial Intelligence, Low-Field MRI, Open dataset
We release a new raw k-space dataset M4Raw acquired by the low-field MRI. Currently, it contains multi-channel brain data of 180 subjects each with 18 slices x 3 contrasts (T1w, T2w, and FLAIR). Moreover, each contrast consists of two or three repetitions (a.k.a. NEXs), leading to more than 25k trainable slices in total, which can be used in various ways by the low-field MRI community. It can be accessed via http://github.com/mylyu/M4Raw.
Introduction
Low-field MRI has been revisited recently for better MRI accessibility and affordability worldwide. The key limiting factor at low field is the SNR per unit time, to which new advances in data-driven methods may provide promising solutions. However, there is a lack of publicly available low-field MRI datasets that can support machine learning training and cross-study comparison, as fastMRI1 and Calgary-Campinas-3592 do for the high-field MRI research community.To fill the gap, we release a new raw k-space dataset acquired by the low-field MRI. Currently, it contains multi-channel brain data of 180 subjects each with 18 slices x 3 contrasts (T1w, T2w, and FLAIR). Moreover, each contrast consists of two or three repetitions (a.k.a. NEXs), leading to more than 25k trainable slices in total, which can be used in various ways by the low-field MRI community.
We name this dataset M4Raw (Multi-contrast Multi-repetition Multi-channel Raw k-space dataset for low-field MRI reconstruction). It can be accessed via http://github.com/mylyu/M4Raw. In this abstract, we demonstrate its potential usage in parallel imaging reconstruction and denoising tasks. Moreover, it could also be used for other tasks such as (motion) artifact correction and super-resolution.
Methods
Imaging protocolWith approval by the local Institutional Review Board, 180 healthy volunteers have been enrolled with written informed consents. The majority of the subjects were college students. For each subject, axial brain MRI data were acquired on a permanent magnet based 0.3T whole-body scanner (Oper-0.3, Ningbo Xingaoyi) using a four-channel head coil. Three sequences were acquired with the parameters listed below:
| | Type | Matrix size | TR/TE | Scan time per repetition | Echo train length | Echo spacing | No. of repetitions | Common parameters | |
| T1w | SE | 256x195 | 500/18.4 | 1min 38s | 1 | N/A | 3 | FOV=240mmx240mm; No. of slices=18; Slice thickness=5mm; Slice gap=1mm; Sampling bandwidth =31.25KHz. | |
| T2w | FSE | 256x195 | 5500/128 | 1min 23s | 13 | 16.0ms | 3 | ||
| FLAIR | IR-FSE | 256x198 | 7500/98, TI =1655 | 2min 15s | 11 | 16.3ms | 2 | ||
Processing and data format
Individual repetitions were saved and raw k-space data were exported and converted to HDF5 files with ISMRMRD-compatible headers3. The k-space data were multiplied by a phase ramp for off-center correction according to the coordinates in the DICOM header. Data were split into a training subset of 144 subjects and a validation subset of 36 subjects randomly without overlap.
Usage demonstration
Parallel imaging reconstruction
We trained an end-to-end VarNet4 model on M4Raw for the demonstration of parallel imaging reconstruction. Data were Cartesian undersampled at SENSE factor = 2 and 3. Its performance was evaluated on the single repetition data in the validation subset and compared with the classical GRAPPA algorithm5.
Denoising
We trained a NAFNet model 6(current state-of-the-art natural image restoration model) and a ResUNet7 model on M4Raw for the demonstration of denoising. Single repetition data were used as inputs, and Multi-repetition averaged data were used as labels. The models were compared with classical BM3D algorithms and PSNR/SSIM values were computed for quantitative metrics.
Quality control
As subject motion might occur between scans, we calculated the PSNR for each repetition with regard to the first repetition as a measure of motion severity.
Results
As shown in Figure 1, the four channels have much less sensitivity variation compared with typical high-field coils, mainly due to the coil design at low field8 and long EM wavelength. However, there were still obvious in-plane phase differences among coils. This created the possibility of parallel imaging acceleration as validated in Figure 2, where the machine learning method VarNet4 resulted in greatly improved image quality over GRAPPA. Even at R=3, VarNet had acceptable image quality with only slight artifacts along the undersampled direction (left-right direction). For denoising, machine learning methods (NAFNet and ResUNet) yielded similar results, both better than BM3D which was over-smoothed, as shown in Figure 3.Figure 4 plots the distribution of PSNRs of each repetition with regard to the first repetition. Most of the scans had PSNR over 26, above which the motion could be considered minor according to our visual inspection. Note that we nevertheless release all the data to reflect the challenges in real MRI acquisitions; the M4Raw users can determine a study-specific criterion for motion correction or data rejection if needed.
Discussion and Conclusions
We release a dataset M4Raw publicly available for low-field MRI reconstruction. This dataset enables flexible training of machine learning-based reconstruction models due to the multi-contrast multi-repetition raw data acquisitions. It also provides a reproducible benchmark for fair comparisons across different studies.This open dataset not only can serve low-field research but also can be used as an external validation for general reconstruction algorithms. We plan to cover more organs such as the spine and knee in future versions, and possibly include other contrasts such as DWI.
Acknowledgements
This study is supported in part by Natural Science Foundation of Top Talent of Shenzhen Technology University (Grants No. 20200208 to Lyu, Mengye) and the National Natural Science Foundation of China (Grant No. 62101348 to Lyu, Mengye)References
1. Zbontar J, Knoll F, Sriram A, et al. fastMRI: An Open Dataset and Benchmarks for Accelerated MRI. ArXiv181108839 Phys Stat. Published online December 11, 2019. Accessed September 27, 2020. http://arxiv.org/abs/1811.08839
2. Souza R, Lucena O, Garrafa J, et al. An open, multi-vendor, multi-field-strength brain MR dataset and analysis of publicly available skull stripping methods agreement. NeuroImage. 2018;170:482-494. doi:10.1016/j.neuroimage.2017.08.021
3. ISMRM Raw data format: A proposed standard for MRI raw datasets - Inati - 2017 - Magnetic Resonance in Medicine - Wiley Online Library. Accessed November 8, 2022. https://onlinelibrary.wiley.com/doi/10.1002/mrm.26089
4. Sriram A, Zbontar J, Murrell T, et al. End-to-End Variational Networks for Accelerated MRI Reconstruction. Published online April 15, 2020. doi:10.48550/arXiv.2004.06688
5. Griswold MA, Jakob PM, Heidemann RM, et al. Generalized autocalibrating partially parallel acquisitions (GRAPPA). Magn Reson Med. 2002;47(6):1202-1210. doi:10.1002/mrm.10171
6. Chen L, Chu X, Zhang X, Sun J. Simple Baselines for Image Restoration. Published online August 1, 2022. doi:10.48550/arXiv.2204.04676
7. Zhang K, Li Y, Zuo W, Zhang L, Van Gool L, Timofte R. Plug-and-Play Image Restoration with Deep Denoiser Prior. IEEE Trans Pattern Anal Mach Intell. Published online 2021.
8. Marques JP, Simonis FFJ, Webb AG. Low-field MRI: An MR physics perspective. J Magn Reson Imaging. 2019;49(6):1528-1542. doi:10.1002/jmri.26637
Figures
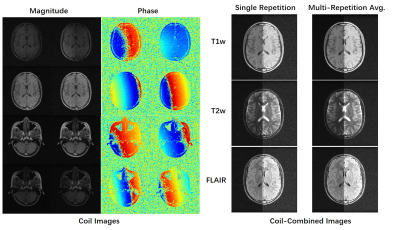
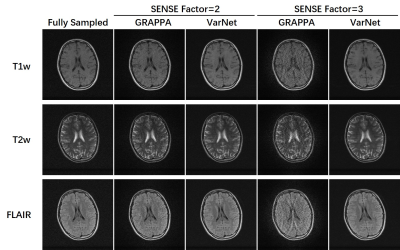
Figure 2: Demonstration of training parallel imaging models on the M4Raw dataset. Despite the relatively small number of coils, it was feasible to study low to moderate acceleration up to 3-fold.
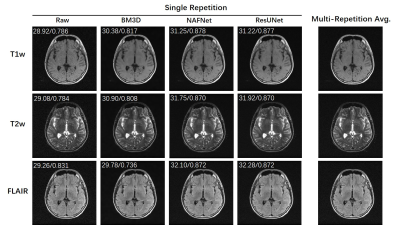
Figure 3: Demonstration of training denoising models on the M4Raw dataset. Because multi-repetition data were separately saved, it was possible to train/evaluate a wide range of denoising models very flexibly. The PSNR/SSIM values are labeled on the top of each image using multi-repetition averaged images as the reference.
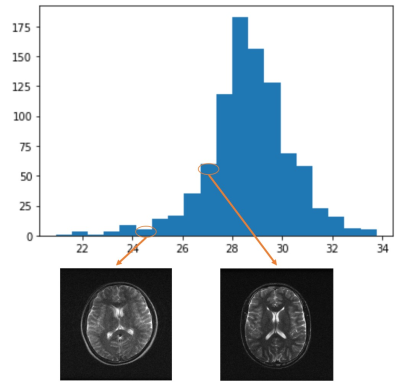
Figure 4: Histogram of PSNR of each repetition (taking the first repetition as reference) to detect possible subject motion between scans. The majority of scans had PSNR values higher than 26, above which no apparent blurring was observed on the repetition-averaged images. To reflect challenges in real MRI acquisitions, we release all the data without setting an absolute threshold for exclusion. The M4Raw users can determine a study-specific criterion for motion correction and data rejection if needed.