0403
Developing an Open Access Brain Metastasis Database: Yale Brain Metastasis Database1Department of Radiology, Yale School of Medicine, New Haven, CT, United States, 2University of Essen, Essen, Germany, 3Ludwig Maximilians Universität (LMU), Munich, Germany, 4University of Goettingen, Goettingen, Germany, 5Visage Imaging, Dusseldorf, Germany, 6Department of Radiology and Biomedical Imaging, Yale University, New Haven, CT, United States, 7University of Ulm, Ulm, Germany, 8University of Leipzig, Leipzig, Germany
Synopsis
Keywords: Tumors, Machine Learning/Artificial Intelligence, Brain Metastasis
While there are many machine learning (ML) algorithms for brain metastasis (BM) detection and segmentation, very few have been validated on external datasets. There is a critical need for open access BM datasets for development and validation of more robust algorithms. Here, we present the Yale Brain Metastasis database of 290 patients with annotated segmentations of BM on T1 post-gadolinium and associated survival information. A subset of 228 patients have FLAIR segmentations, clinical features, and qualitative imaging features. Open access of this database will greatly aid in the development and validation of new AI algorithms for BM detection and segmentation.INTRODUCTION
Brain metastases (BM) are the most common brain tumor in adults although there is a paucity of available BM datasets for open access research. While several machine learning (ML) algorithms have been developed to classify and characterize BM, very few studies have performed external validation of their algorithms due to difficulty in obtaining secure and anonymized datasets from outside institutions for external validation1,2. Furthermore, there are no US FDA-cleared AI algorithms to date with BM focus3.Additionally, the images generally used in the literature for algorithm training are highly curated and not representative of those encountered in clinical practice. One potential solution to building more robust algorithms for BM using more representative images is to make institutional datasets publicly available. Here, we present the Yale Brain Metastasis database that includes annotated images of BM with associated clinical outcome information.
METHODS
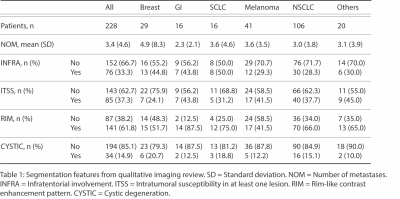
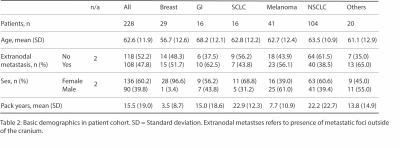
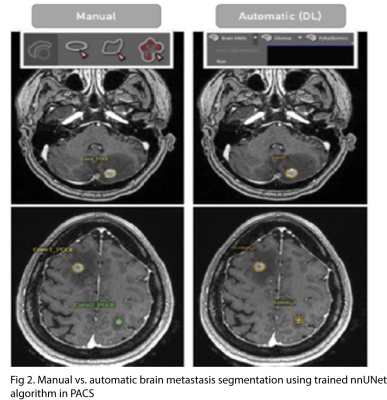
Our database includes 290 patients (1133 brain metastases) with T1 post-contrast images and corresponding survival data. Volumetric segmentations of enhancing tumor and peritumoral edema were generated in a semiautomatic workflow using nnUNet algorithm4 trained on an excluded subset. Segmentations were validated by a board-certified neuroradiologist.A subset of 228 patients from the database also had FLAIR images and information about qualitative imaging features (number of metastases, infratentorial involvement, intratumoral susceptibility in at least one lesion, rim-like contrast enhancement pattern, and cystic degeneration) and associated clinical features (age, sex, presence of extranodal metastasis, and smoking pack-year history).
RESULTS
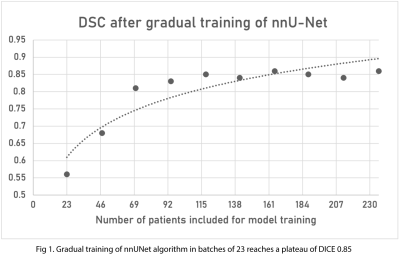
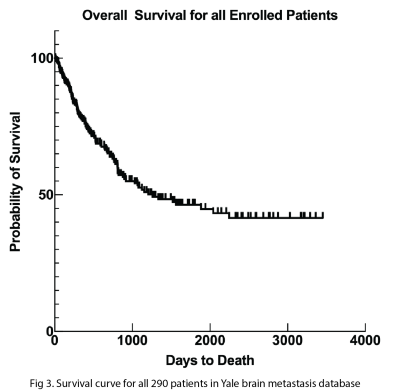
Gradual training of nnUNet was performed using batches of 23 patients. DSC scores for segmentation of the contrast enhancing portion of the tumor reached a plateau of DICE 0.85 after 4 batches. 100% of the brain metastases had segmentation of peritumoral edema on FLAIR.The median overall survival for all 290 patients in our database was 3.4 years. The overall 1-year survival rate was around 77%, and the overall 5-year survival rate was around 45%.
Of the 228 patients who had associated imaging and clinical outcome information available, the following was the breakdown of origin of brain metastases: breast (29), GI (16), SCLC (16), melanoma (41), NSCLC (106), and others (20). The mean number of metastases among all cancer origins was 3.4 per patient with the following breakdown by origin subtype: breast (4.9), GI (2.3), SCLC (3.6), melanoma (3.6), NSCLC (3.0), and others (3.1). 33.3% of all patients had infratentorial involvement with the following breakdown by origin: breast (44.8%), GI (43.8%), SCLC (50%), melanoma (29.3%), NSCLC (28.3%), and others (30%). 37.3% of all patients had intratumor susceptibility in at least one lesion with the following breakdown by origin: breast (24.1%), GI (43.8%), SCLC (31.2%), melanoma (41.5%), NSCLC (37.7%), and others (45%). 61.8% of all patients had rim-like contrast enhancement pattern with the following breakdown: breast (51.7%), GI (87.5%), SCLC (75%), melanoma (41.5%), NSCLC (66%), and others (65%). 14.9% of all patients had cystic degeneration with the following breakdown: breast (20.7%), GI (12.5%), SCLC (18.8%), melanoma (12.2%), NSCLC (15.1%), and others (10%).
The mean age of patients within the subset of 228 patients was 62.6 years with 60.2% female and 39.8% male. 47.8% of patients had extranodal metastasis with the following breakdown: breast (51.7%), GI (62.5%), SCLC (43.8%), melanoma (56.1%), NSCLC (38.5%), and others (65%). The mean pack-year smoking history for all patients was 15.5 with the following breakdown: breast (3.5), GI (15), SCLC (22.9), melanoma (7.7), NSCLC (22.2), and others (13.8).
DISCUSSION
Our dataset includes a large cohort of patients with associated radiographic and clinical information. It was generated in a streamlined approach involving direct clinical production to research server database using a forward-based hash to maintain a deidentified patient jacket. AI-based BM detection and segmentation allows radiologists to revise segmentation masks and batch export images/segmentations into NIfTI for sharing.There are few initiatives aimed at providing open access datasets of brain metastases. The Cancer Imaging Archive (TCIA) is one initiative that builds publicly available imaging datasets but primarily includes GBM, low-grade, and high-grade gliomas5. The Stanford Center for Artificial Intelligence in Medicine & Imaging’s BrainMetShare initiative has published an open access dataset of 156 MRI studies with ground-truth BM segmentations by neuroradiologists on pre- and post-contrast whole brain images6. The dataset includes brain metastases with primary origin from lung (n = 99), breast (n = 33), melanoma (n = 7), genitourinary (n = 7), GI (n = 5), and others (n = 5). Our dataset consists of more patients with a larger number of lung metastases (122) broken down by lung cancer subtype (NSCLC vs. SCLC). In addition, our dataset provides more information about imaging features, such as rim-enhancement pattern and cystic degeneration, as well as more patient clinical information, such as overall survival, smoking history, and presence of extranodal metastasis.
CONCLUSION
We demonstrate a novel dataset that consists of 290 patients (1133 brain metastases) that underwent individual segmentations on post-contrast MRI as well as a subset of 228 patients with FLAIR images, additional qualitative imaging features, and clinical information. Availability of this data on an open access environment will provide the critically needed data for novel algorithm development of brain metastases.Acknowledgements
No acknowledgement found.References
1. Jekel L, Brim WR, von Reppert M, et al. Machine Learning Applications for Differentiation of Glioma from Brain Metastasis-A Systematic Review. Cancers. 2022;14(6):1369. doi:10.3390/cancers14061369
2. Cho SJ, Sunwoo L, Baik SH, Bae YJ, Choi BS, Kim JH. Brain metastasis detection using machine learning: a systematic review and meta-analysis. Neuro-Oncol. 2021;23(2):214-225. doi:10.1093/neuonc/noaa232
3. New ACR DSI Searchable FDA-Cleared Algorithm Catalog Can Ease Medical Imaging AI Integration. Accessed November 9, 2022. https://www.acrdsi.org/News-and-Events/New-ACR-DSI-Searchable-FDA-Cleared-Algorithm-Catalog-Can-Ease-Medical-Imaging-AI-Integration
4. Isensee F, Jaeger PF, Kohl SAA, Petersen J, Maier-Hein KH. nnU-Net: a self-configuring method for deep learning-based biomedical image segmentation. Nat Methods. 2021;18(2):203-211. doi:10.1038/s41592-020-01008-z
5. The Cancer Imaging Archive (TCIA): Maintaining and Operating a Public Information Repository | SpringerLink. Accessed November 9, 2022. https://link.springer.com/article/10.1007/s10278-013-9622-7
6. Liew A, Lee CC, Subramaniam V, Lan BL, Tan M. Gradual Self-Training via Confidence and Volume Based Domain Adaptation for Multi Dataset Deep Learning-Based Brain Metastases Detection Using Nonlocal Networks on MRI Images. J Magn Reson Imaging JMRI. Published online October 8, 2022. doi:10.1002/jmri.28456