0401
A Remote Sequence Streaming Interface for Open Innovation
Thomas Kluge1 and Christoph Forman1
1Siemens Healthcare GmbH, Erlangen, Germany
1Siemens Healthcare GmbH, Erlangen, Germany
Synopsis
Keywords: Data Acquisition, Pulse Sequence Design, Open Innovation; Reproducible Research
Open innovation and reproducible research have become increasingly important in recent years. Furthermore, software version dependent application development introduces significant efforts to developers and researchers in maintaining different versions of applications for a variety of scanner software releases. In this work, we propose a software-version and programming language independent interface for data acquisition on Siemens Healthcare MAGNETOM scanners. This is achieved by generic interfaces and streaming protocols. A wide range of development and deployment opportunities is enabled with a web-API. The use of the interface is exemplarily shown with a prototype FLASH sequence and demonstrated with a phantom scan.Background
In recent years, open innovation and reproducible research have become increasingly important in the research community. For image reconstruction, significant efforts have been spent to establish a vendor-independent standard for the storage of MRI raw data (ISMRMRD1). Reconstruction algorithms supporting this format can be distributed enabling reproduction of results and application to new data. This can be directly performed offline for research purposes, while its use within the clinical workflow at the scanner may require integration into the MR system software2. For data acquisition, several MR sequence development environments have been proposed3-5. While these environments claim vendor-independence, the integration on the scanner relies on a vendor specific adapter module provided with these environments. This module is deeply integrated into the vendor’s scanner control software and must be adapted and re-released for each software update. This creates significant efforts for the developers and users of such an environment. To address these shortcomings and to fully support reproducible research, in this work we propose a software-version and programming language independent interface for data acquisition on Siemens Healthcare MAGNETOM scanners. The use of the interface is exemplarily shown with a prototype FLASH sequence and demonstrated with a phantom scan.Methods
The structure of the interface is shown in Figures 1 and 2. The Remote Sequence Streaming Interface has a central role connecting the vendor control software to a Remote Sequence via a plug-in mechanism. The interface allows exchange of data between scanner and Remote Sequence using streaming protocols. As shown in Figure 3 the scanner control data is streamed as a series of commands that each are identified by a unique identifier (UUID) and remain stable across all software versions. With this design, new commands can be added without modifying the interface or rendering existing Remote Sequences invalid. While scanner control data is sent to the scanner, the measured data is returned to the Remote Sequence. Additionally, the interface features optional methods to parameterize the Remote Sequence using the vendor’s user interface. While the interface can be directly implemented in C++, adapter modules are provided to physically decouple the Remote Sequence from the scanner and to connect to the control software via a web-API using JSON. This web-API enables various deployment options for the Remote Sequence and the use of multiple programming languages and, as of now, control data types used in the commands are available in C++, C#, Matlab, and Python. A prototype software was developed and fully integrated in the scanner control software of a 3T clinical MR scanner (MAGNETOM Vida, Siemens Healthcare, Germany). A FLASH sequence was developed as Remote Sequence utilizing the commands provided by the interface. This sequence was validated against the original sequence directly implemented in the vendor’s sequence development environment. Phantom experiments were performed with both sequences and following parameters: TE=10ms, TR=20ms, flip-angle=15deg, FOV=256x256mm2, pixel size=1.0x1.0mm2, and a slice thickness of 5.0mm. A graphical tool relating the stream of commands to the resulting MR-sequence has been implemented and is shown in Figure 4.Results
The prototype FLASH sequence and various deployment options successfully have been validated against the vendor's original implementation utilizing a sequence-diagram-based validation toolkit introduced earlier6. The scanner integration was successful, and images acquired with the prototype FLASH sequence show no visible differences to the original, see Figure 5.Discussion
This study showed that it is feasible to develop and run sequences with the proposed interface. With about 10 classes and 20 methods the C++ interface should enable an easy familiarization and support the development of sequence applications. The streamed commands reflect the scanners capabilities in an abstract manner and a single command can easily be understood and utilized. Currently the interface is limited to static sequences that can be completely precomputed before execution. Future work includes its extension with commands that interact with physiological signals from patients during runtime and, hence, enable more advanced applications. While the command set easily can be upgraded, the structure of single commands aims at long term stability. For example, hardware support for an arbitrary number of channels for field generation is already foreseen. The current prototype showed no noticeable delays when using the user interface (UI) or during data acquisition. For the operator, the full integration into the vendor’s UI provided identical usability as a conventional sequence even though the sequence application logic was provided via web interface. For researchers and developers, the use of the web-API extends the development opportunities to multiple programming languages and environments. Furthermore, the employed web technology enables new deployment options for applications via network connection or virtualized containers.Conclusion
The proposed interface provides a standardized sequence development across different software versions. A wide range of development and deployment opportunities is enabled with the web-API. Combining the proposed interface with other established sequence development environments is desirable to further support open innovation and reproducible research. Future work should integrate the ISMRMRD format leading to the development of a full application combining both sequence and reconstruction interfaces.Acknowledgements
No acknowledgement found.References
- Inati SJ, et al. ISMRM Raw data format: A proposed standard for MRI raw datasets. Magn. Reson. Med., 77: 411-421. https://doi.org/10.1002/mrm.26089
- Chow K, et al. Prototyping Image Reconstruction and Analysis with FIRE. In: proc. SCMR Virtual Scientific Sessions 2021, p. 838972
- Santos JM, et al. Flexible real-time magnetic resonance imaging framework. In: proc. IEMBS, 2004, pp. 1048-1051, https://doi.org/10.1109/IEMBS.2004.1403343
- Cordes C, et al. Portable and platform-independent MR pulse sequence programs. Magn. Reson. Med., 2020, 83: 1277–1290. https://doi.org/10.1002/mrm.28020
- Layton KJ, et al. Pulseq: A rapid and hardware-independent pulse sequence prototyping framework. Magn. Reson. Med., 2017, 77: 1544-1552
- Kluge T, et al. A Comprehensive MR Fingerprinting Development Kit (MRFDK) In: proc. ISMRM, 2022, p. 4798
Figures
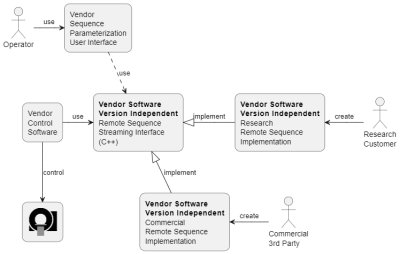
The C++ remote sequence streaming interface is used by the vendor's control software and optionally by the vendor's sequence parametrization GUI. This interface is carefully designed and versioned and independent of the vendor software, i.e., the vendor software can evolve while this remote sequence streaming interface does not change. The MR sequences implemented by research customers as well as by commercial 3rd-parties can be executed unmodified with future vendor software versions.
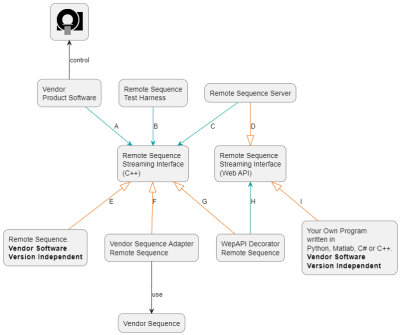
Overview of the C++ remote sequence streaming interface, its equivalent web version, and the software components using (green --> arrows) or implementing/providing (orange --|> arrows) the interfaces. The letters attached to the arrows allow for describing potential deployment configurations. E.g., A-E: C++ remote sequence implementation is directly loaded by the product software. E.g., A-G-H-D-C-E: C++ remote sequence implementation is completely decoupled from the vendor product software via the web API.
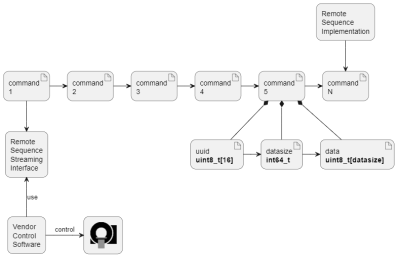
he MR-sequence running on the vendor's scanner is defined by the remote sequence via a stream of commands. As shown for the command 5, each command has the same basic structure, i.e., a UUID identifying the command, the size of the data associated with the command and the command content data itself. In the remote sequence streaming interface, one single method is required for streaming the commands and this method does not have to be changed when new commands are added to the set of supported commands.
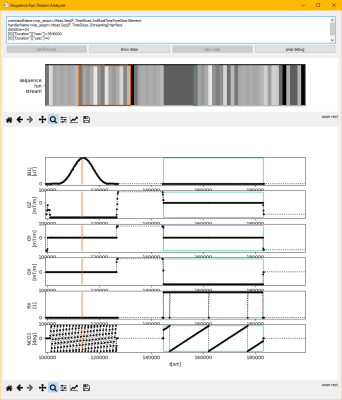
A demonstrator GUI for interactively analyzing a remote sequence control data stream. The vertically striped plot in the middle shows the series/stream of commands defining the sequence. The multi-plot below shows the resulting MR-sequence diagrams. A command in the control data stream can be marked in green and the affected time period in the MR-sequence is highlighted. A time point in the MR-sequence can be marked in orange and the commands affecting that time are marked in the control data stream. The text-box on the top shows the content of the command marked in green.
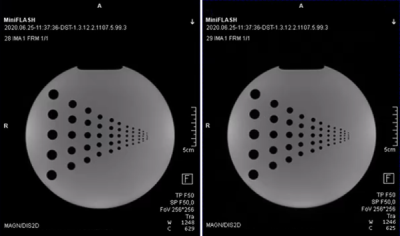
The resulting images of the phantom scan. The left image shows the reconstructed image of the FLASH sequence directly implemented in the vendor’s software development environment. The right image shows the reconstructed image of the prototype FLASH sequence developed as Remote Sequence utilizing the commands provided by the interface. Both images were reconstructed using the same image reconstruction pipeline at the scanner.
DOI: https://doi.org/10.58530/2023/0401