0398
Cross-vendor three-dimensional multiparametric mapping of the human brain: A traveling-subject and patient study
Shohei Fujita1,2, Borjan Gagoski3,4, Ken-Pin Hwang5, Marcel Warntjes6,7, Kazumasa Yokoyama8,9, Issei Fukunaga1, Wataru Uchida1, Yuya Saito1, Rina Tachibana1, Tomoya Muroi1, Toshiya Akatsu1, Akihiro Kasahara2, Ryo Sato2, Tsuyoshi Ueyama2, Christina Andica1, Akifumi Hagiwara1, Koji Kamagata1, Shiori Amemiya2, Hidemasa Takao2, Nobutaka Hattori9, Osamu Abe2, and Shigeki Aoki1
1Department of Radiology, Juntendo University, Tokyo, Japan, 2Department of Radiology, The University of Tokyo, Tokyo, Japan, 3Fetal Neonatal Neuroimaging and Developmental Science Center, Boston Children's Hospital, Boston, MA, United States, 4Department of Radiology, Harvard Medical School, Boston, MA, United States, 5Department of Imaging Physics, MD Anderson Cancer Center, Houston, TX, United States, 6SyntheticMR, Linköping, Sweden, 7Center for Medical Imaging Science and Visualization (CMIV), Linköping University, Linköping, Sweden, 8Tousei center for neurological diseases, Shizuoka, Japan, 9Department of Neurology, Juntendo University, Tokyo, Japan
1Department of Radiology, Juntendo University, Tokyo, Japan, 2Department of Radiology, The University of Tokyo, Tokyo, Japan, 3Fetal Neonatal Neuroimaging and Developmental Science Center, Boston Children's Hospital, Boston, MA, United States, 4Department of Radiology, Harvard Medical School, Boston, MA, United States, 5Department of Imaging Physics, MD Anderson Cancer Center, Houston, TX, United States, 6SyntheticMR, Linköping, Sweden, 7Center for Medical Imaging Science and Visualization (CMIV), Linköping University, Linköping, Sweden, 8Tousei center for neurological diseases, Shizuoka, Japan, 9Department of Neurology, Juntendo University, Tokyo, Japan
Synopsis
Keywords: Quantitative Imaging, Precision & Accuracy, Cross-vendor
Multiparametric techniques compatible with multiple vendors to facilitate the pooling of data among different sites and vendors are desired. Here, we developed a vendor-standardized whole-brain multiparametric mapping scheme based on 3D-QALAS. Intra-scanner repeatability and inter-vendor reproducibility were evaluated on test-retest session data on five different 3T systems from four MRI vendors (GE, Philips, Siemens, and Canon). T1 and T2 relaxation times and proton density values derived from 3D-QALAS showed coefficient of variations of <4.0% across scanners from different vendors. Finally, we performed an inter-vendor validation on multiple sclerosis patients to assess the feasibility of the scheme in real-world clinical settings.Introduction
Multiparametric mapping enables the simultaneous quantification of tissue parameters, which could be used to characterize normal distributions in healthy individuals and thus be used to objectively and sensitively detect neurological diseases.1-5 A multiparametric technique compatible with multiple vendors is necessary to pool data among different sites and vendors to increase the size and diversity of the applicable patient population.6,7 However, few technologies have shown compatibility across vendors or provided reproducible values across MRI scanners from different vendors.One cross-vendor multiparametric technique is 3D-quantification using an interleaved Look–Locker acquisition sequence with a T2 preparation pulse (3D-QALAS).4,8 Here, we developed a vendor-standardized whole-brain multiparametric mapping scheme based on 3D-QALAS and assessed the intra-scanner repeatability and inter-vendor reproducibility of T1, T2, and proton density (PD) values in five different 3T systems from four major MRI vendors. Further, we performed an inter-vendor validation on patients with multiple sclerosis to assess the feasibility of 3D-QALAS in real-world clinical settings.
Methods
Acquisition protocolOur prospective multi-institutional study was approved by the local institutional review board of each institution. Participants provided written informed consent. Five 3T scanners manufactured by four vendors (GE Healthcare, Philips Healthcare, Siemens Healthcare, and Canon Medical Systems) were included from two academic institutions. The 3D-QALAS imaging parameters were kept as close as possible across vendors (Table 1).
Data Acquisition
Ten healthy volunteers (4 women; mean age ± SD, 28.0 ± 5.6 years) underwent test-retest of vendor-standardized 3D-QALAS sequence with each scanner. Subjects were repositioned between the acquisitions to assess variability due to subject positioning. Each participant underwent imaging on all the scanners within two months. To further assess the feasibility of 3D-QALAS in real-world clinical settings, nine female patients with multiple sclerosis (mean age ± SD, 28.0 ± 5.6 years; median Expanded Disability Status Scale at imaging, 1.5; disease duration, 5.2 ± 4.0 years) underwent imaging on two scanners from different vendors. Retest was not feasible for patients.
Postprocessing
Whole-brain T1, T2, and PD map were derived via voxel-wise signal modeling-based parameter fitting on the 3D-QALAS source images using prototype SyMRI software. FreeSurfer9 was used to perform brain region-wise analysis with the Desikan-Killiany atlas.10 The mean T1, T2, and PD values were calculated for each brain structure in cortical gray matter (GM), subcortical GM, and white matter (WM) structures and WM lesions in patients.
Assessment
Intra-scanner repeatability for each scanner and structure was assessed using the within-subject coefficient of variation (CV), defined as the standard deviation [SD] of the three quantitative maps derived from test and retest over the mean of both time points.11 Inter-vendor reproducibility was also assessed using within-subject CV, defined as the SD divided by mean value calculated across all five scanners. Linearity, slope, intercept, and limits of agreement (defined as mean ± 1.96 × SD) were used for comparisons among scanners. The intraclass correlation coefficient (ICC) using two-way random-effects model was also calculated to assess agreement between scanners.
Results
Quantitative values of five scanners from four vendorsRepresentative quantitative maps of a healthy subject are shown in Figure 1. The mean intra-scanner CVs for each scanner and structure ranged from 0.5 to 2.1% (Figure 2A). The overall structure-wise test-retest repeatability (mean intra-scanner CVs of the five scanners) were 1.6%, 1.1%, and 0.7% for T1, T2, and PD, respectively (Figure 2B). Overall, a high inter-vendor reproducibility was observed for all parameter maps (Figure 3A and 3B, ICC = 0.99, 0.97, and 0.98 for T1. T2, and PD, respectively). The structure-wise inter-scanner CV values are shown in Figure 3C.
Inter-vendor patient study
Representative quantitative maps of a patient with multiple sclerosis are shown in Figure 4A. High agreement was observed among the scanners for all structure measurements, including WM lesions (Figure 4B to 4D, ICC = 0.98, 0.92, and 0.94 for T1. T2, and PD, respectively).
Discussion
In this study, we examined the intra-scanner and inter-vendor reproducibility of quantitative measurements obtained with 3D-QALAS, a cross-vendor whole-brain multiparametric mapping technique. Although the number of participants was small, this study included scanners from four major vendors, covering more than 90% of the global install base. Our results show that the T1, T2, and PD values measured with 3D-QALAS were comparable across scanners and vendors. Further, applying vendor-specific advanced acceleration techniques may lead to greater adoption in research and clinical settings.12Importantly, the results demonstrate that the reproducibility coefficient (defined as 2.77 × within-subject CV) was smaller than the difference between the quantitative values of the lesion and the normal tissue: this indicates that lesions can be distinguished from normal tissue even when data from scanners from multiple vendors are pooled.
We did not account for B1 inhomogeneity in this study, which may introduce bias and variance in the quantitative maps, especially the T1 maps.13 Integrating B1 correction into the framework will be conducted.
Conclusion
In a cohort consisting of healthy volunteers and patients, T1, T2, and PD values measured with 3D-QALAS were in high agreement across 3T scanners from four major MRI vendors. This validation study may serve as a base for larger multi-institutional studies.Acknowledgements
No acknowledgement found.References
- Jara H, Sakai O, Farrher E, et al. Primary Multiparametric Quantitative Brain MRI: State-of-the-Art Relaxometric and Proton Density Mapping Techniques. Radiology. 2022;305(1):5-18. doi:10.1148/radiol.211519
- Ma D, Gulani V, Seiberlich N, et al. Magnetic resonance fingerprinting. Nature. 2013;495(7440):187-192. doi:10.1038/nature11971
- Wang F, Dong Z, Reese TG, et al. Echo planar time-resolved imaging (EPTI). Magn Reson Med. 2019;81(6):3599-3615. doi:10.1002/mrm.27673
- Fujita S, Hagiwara A, Hori M, et al. Three-dimensional high-resolution simultaneous quantitative mapping of the whole brain with 3D-QALAS: An accuracy and repeatability study. Magn Reson Imaging. 2019;63:235-243.
- Ma S, Nguyen CT, Han F, et al. Three-dimensional simultaneous brain T1 , T2 , and ADC mapping with MR Multitasking. Magn Reson Med. 2020;84(1):72-88. doi:10.1002/mrm.28092
- Seiberlich N, Gulani V, Campbell-Washburn A et al. Quantitative Magnetic Resonance Imaging, Volume 1 – 1st Edition. 2020.
- QIBA Concepts. https://qibawiki.rsna.org/index.php/QIBA_Concepts
- Kvernby S, Warntjes MJ, Haraldsson H, Carlhäll CJ, Engvall J, Ebbers T. J Cardiovasc Magn Reson. 2014 Dec 20;16:102. Simultaneous three-dimensional myocardial T1 and T2 mapping in one breath hold with 3D-QALAS.
- Fischl B. FreeSurfer. Neuroimage 2012;62:774–781
- 10. Desikan RS, Segonne F, Fischl B, et al. An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. Neuroimage 2006;31:968–980.
- Weingärtner S, Desmond KL, Obuchowski NA, et al. Development, validation, qualification, and dissemination of quantitative MR methods: Overview and recommendations by the ISMRM quantitative MR study group. Magn Reson Med. 2022;87(3):1184-1206.
- Cho J, Gagoski B, Kim T, et al. Wave-Encoded Model-based Deep Learning for Highly Accelerated Imaging with Joint Reconstruction. arXiv. 2022; 2202.02814
- Boudreau M, Tardif CL, Stikov N, Sled JG, Lee W, Pike GB. B1 mapping for bias-correction in quantitative T1 imaging of the brain at 3T using standard pulse sequences. J Magn Reson Imaging. 2017;46(6):1673-1682. doi:10.1002/jmri.25692
Figures
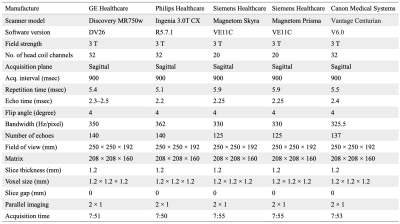
Table 1. Standardized sequence parameters of 3D-QALAS. The 3D-QALAS imaging parameters were kept as close as possible across vendors, and a minimal acceleration technique was applied to make a head-to-head comparison. 3D-QALAS, 3D-quantification using an interleaved Look–Locker acquisition sequence with a T2 preparation pulse.
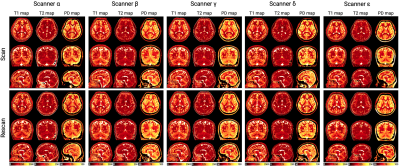
Figure 1. Representative T1, T2, and proton density (PD) maps from a healthy traveling subject. Each traveling subject underwent a test-retest session on each of the five 3T MRI scanners from four vendors (i.e., GE Healthcare, Philips Healthcare, Siemens Healthcare, Canon Medical Systems). Subjects were repositioned between the acquisitions. The retest maps were aligned to the initial maps for visualization.
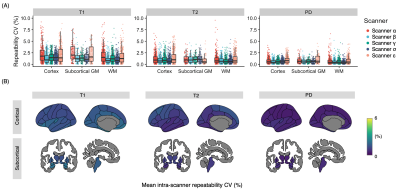
Figure 2. Intra-scanner repeatability. (A) Boxplot showing structure-wise coefficient of variation (CV) for each scanner. Each dot represents a brain region on the Desikan-Killiany atlas. (B) Region-wise intra-scanner repeatability. For each brain structure, the intra-scanner CV value of the five scanners was calculated for each subject, and the mean value across subjects was displayed on schematic brain illustrations.
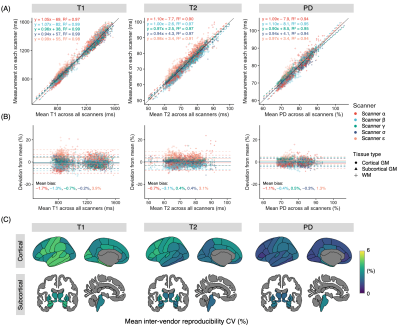
Figure 3. (A) Scatter and Bland-Altman plots showing inter-scanner and inter-vendor agreement. Each dot represents a brain region. The solid line is the identity line. The dashed line is the linear fit for each scanner. (B) Bland–Altman plots showing inter-scanner biases. (C) Region-wise inter-scanner CVs for each structure calculated using the 1st scan from each scanner. CV, coefficient of variation; GM, gray matter; PD, proton density; WM, white matter.
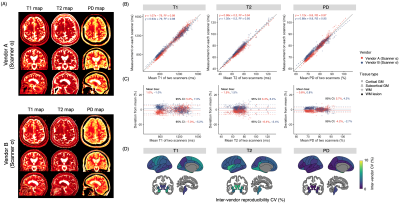
Figure 4. Inter-vendor evaluation on multiple sclerosis patients. (A) Representative maps using two scanners from different manufacturers. (B) Scatter and Bland-Altman plots showing inter-vendor agreement. Each dot represents a brain region. The solid line is the identity line. The dashed line is the linear fit. (C) Bland–Altman plots showing inter-scanner biases. (D) Region-wise inter-scanner CVs. CV, coefficient of variation; GM, gray matter; PD, proton density; WM, white matter.
DOI: https://doi.org/10.58530/2023/0398