0395
Single-shot T2-FLAIR mapping via inversion recovery multiple overlapping-echo acquisition and deep neural network reconstruction1Department of Electronic Science, Fujian Provincial Key Laboratory of Plasma and Magnetic Resonance, Xiamen University, Xiamen, China, 2Department of Magnetic Resonance Imaging, The First Affiliated Hospital of Zhengzhou University, Zhengzhou, China
Synopsis
Keywords: Quantitative Imaging, Quantitative Imaging
T2-weighted imaging via conventional FLAIR sequence can suppress the signal of cerebrospinal fluid (CSF), making it easier for identifying long T2 lesions in the vicinity of the CSF. But it was usually used for qualitative analysis because of its inevitable time-consuming acquisition. In this study, we applied inversion recovery overlapping-echo acquisition together with deep learning-based reconstruction to achieve ultra-fast T2-FLAIR mapping. In vivo results from a healthy volunteer and two glioma patients demonstrate the good accuracy and robustness of our proposed method.Introduction
In the clinical diagnosis of tumors, several studies have demonstrated the great potential of T2 mapping method.1-3 Similarly, FLAIR imaging has an irreplaceable importance4. However, FLAIR requires particularly long TR due to the presence of inversion time TI, which consumes long time for a single acquisition. T2 mapping requires multiple acquisitions, which is a non-negligible factor for clinical application, leading to discouragement of FLAIR mapping in medical imaging. Here, a rapid method, inversion recovery multiple overlapping-echo detachment (IR-MOLED), was developed for T2-FLAIR mapping.Methods
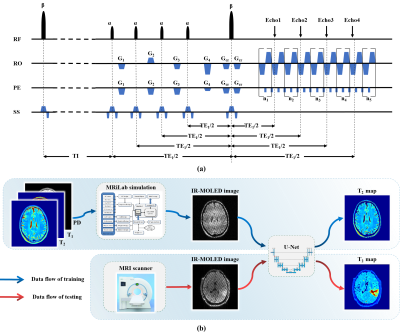
Pulse sequence: Figure 1(a) shows the single-shot IR-MOLED sequence. An inversion pulse was added to the MOLED sequence proposed in our previous work5-7. The value of TI was calculated to eliminate the CSF signal, and then four small-angle excitation pulses were used to acquire echo signals with different TEs at the same k-space.In vivo experiments: In vivo data were acquired from a healthy volunteer and two glioma patients on a 3T MRI scanner (MAGNETOM Prisma, Siemens Healthcare, Erlangen, Germany). The parameters of IR-MOLED were as follows: TI = 2500 ms, TR = 10 s, TE1-TE4 = 22, 52, 82, 110 ms, α = 30° and β = 180°,matrix size = 128×128,slice thickness = 3.5 mm, FOV = 220 × 220 mm2. For comparison, IR-EPI with TI = 2500 ms, TR = 9 s, number of signal averages (NSA) = 6, TE1-TE4 = 63, 80, 95, 110 ms were acquired. Informed consent of each participant was obtained before scans.
Network and training data generation: U-Net was used. 3500 pairs of synthetic images were used for training, and the data of healthy volunteer and glioma patients were used to validate the accuracy and robustness of the method. The pulse sequence and parameters used in simulation were the same as those used in vivo experiments. The multi-contrast images used to produce parametric templates were from the public database IXI (https://brain-development.org/). In the simulation experiment, different from other methods that use software to segment or remove large T2-weighted regions8 to eliminate CSF, we utilized T1 templates in MRiLab9 for signal evolution to obtain original FLAIR data. Then the real and imaginary parts of IR-MOLED images were sent as input to the network, and the output of the network was the corresponding T2 map.
Results
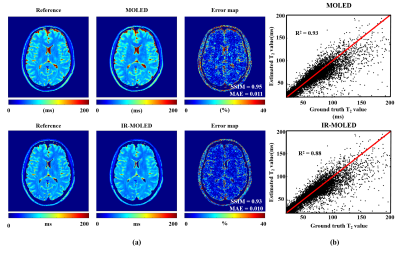
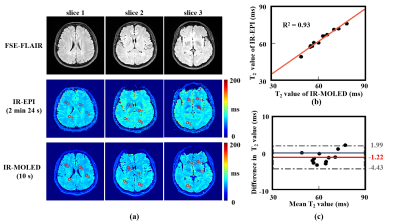
Figure 2 shows the reconstructed maps of numerical brains obtained from MOLED and IR-MOLED. It shows the great performance of MOLED (Structure Similarity (SSIM) = 0.95, Mean Absolute Error (MAE) = 0.011, R2 = 0.93) and IR-MOLED (SSIM = 0.93, MAE = 0.010, R2 = 0.88).Figure 3 illustrates the results of in vivo human brains provided by IR-MOLED, IR-EPI and SE-FLAIR. We can see that our proposed method effectively eliminates the CSF signal. The R2 (0.93) and difference of mean T2 values (-1.22 ms) in ROIs demonstrates high accuracy of IR-MOLED.
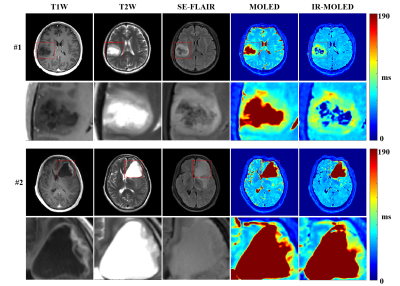
Figure 4 shows the results of two glioma patients. It can be clearly seen that the structure of IR-MOLED and SE-FLAIR sequences are quite consistent in the lesion areas.
Discussion and conclusion
The combination of IR-MOLED acquisition and deep learning-based reconstruction is proven to be effective for glioma T2-FLAIR mapping. Owing to the high temporal resolution of IR-MOLED sequence, ultra-fast T2-FLAIR mapping can be achieved, which offers great possibility for quantitative analysis in clinical diagnosis.Acknowledgements
This work was supported by the National Natural Science Foundation of China under grant numbers 82071913, 22161142024 and U1805261.References
1. Carter JS, Koopmeiners JS, Kuehn-Hajder JE, et al. Quantitative multiparametric MRI of ovarian cancer. Journal of Magnetic Resonance Imaging. 2013;38(6):1501-1509.
2. Liu L, Yin B, Shek K, et al. Role of quantitative analysis of T2 relaxation time in differentiating benign from malignant breast lesions. Journal of International Medical Research. 2018; 46(5):1928-1935.
3. Lee CH, Taupitz M, Asbach P, et al. Clinical utility of combined T2-weighted imaging and T2-mapping in the detection of prostate cancer: A multi-observer study. Quantitative Imaging in Medicine Surgery. 2020;10(9): 1811-1822.
4. Diogo MC, Prayer D, Gruber GM, et al. Echo-planar FLAIR sequence improves subplate visualization in fetal MRI of the brain. Radiology. 2019;292(1):159-169.
5. Zhang J, Wu J, Chen S, et al. Robust single-shot T2 mapping via multiple overlapping-echo acquisition and deep neural network. IEEE Transactions on Medical Imaging. 2019; 38:1801-1811.
6. Yang Q, Lin Y, Wang J, et al. MOdel-Based SyntheTic Data-Driven Learning (MOST-DL): application in single-shot T2 mapping with severe head motion using overlapping-echo acquisition. IEEE Transactions on Medical Imaging. 2022;41(11):3167-3181.
7. Ma L, Wu J, Yang Q, et al. Single-shot multi-parametric mapping based on multiple overlapping-echo detachment (MOLED) imaging. Neuroimage. 2022;263.
8. Kimura T, Yamashita K, Fukatsu K. Synthetic MRI with T2-based water suppression to reduce hyperintense artifacts due to CSF-Partial volume effects in the brain. Magnetic Resonance in Medical Sciences. 2021;20(4):325-337.
9. Liu F, Velikina JV, Block WF, et al. Fast realistic MRI simulations based on generalized multi-pool exchange tissue model. IEEE Transactions on Medical Imaging. 2016; 36:527-537.
Figures