0393
MRI-movienet: fast motion-resolved 4D MRI reconstruction exploiting space-time-coil correlations without k-space data consistency1Department of Medical Physics, Memorial Sloan Kettering Cancer Center, New York, NY, United States, 2Department of Radiology, Memorial Sloan Kettering Cancer Center, New York, NY, United States
Synopsis
Keywords: Machine Learning/Artificial Intelligence, Radiotherapy
Motion-resolved 4D MRI enables free-breathing imaging and access to important physiological information. However, long reconstruction times for 4D MRI techniques like XD-GRASP have restricted routine clinical use. Even with unrolled convolutional networks, reconstruction enforcing data consistency in a high-dimensional space is still long. This work presents a deep learning approach named MRI-movienet that exploits spatial-time-coil correlations without enforcing data consistency to enable 2-fold scan acceleration compared to XD-GRASP and 4D reconstruction in less than 2 seconds. MRI-movienet uses the intrinsic separation into static and dynamic components to avoid hallucinations. MRI-movienet high performance will promote 4D MRI for routine clinical use.INTRODUCTION
Motion-resolved 4D MRI enables free-breathing imaging and access to motion-related physiological information. For example, it can be used to personalize radiotherapy planning of tumors affected by respiratory motion1. However, current state-of-the-art 4D MRI methodology, such as XD-GRASP2, requires relatively long acquisition and particularly long reconstruction times, preventing routine use in clinical studies. Even though deep learning techniques have been applied to reconstruct 4D images3,4, the reconstruction time is still relatively long since they are based on unrolling iterations from compressed sensing and k-space data consistency, which are computationally expensive for the case of 4D imaging and non-Cartesian k-space acquisition.This work presents an alternative deep learning approach for 4D MRI reconstruction named MRI-movienet that exploits space-time-coil correlations in the image domain without enforcing k-space data consistency and the intrinsic separation between static and dynamic components in motion-resolved imaging to avoid hallucinations. MRI-movienet is demonstrated to accelerate the acquisition and reconstruction of XD-GRASP for motion-resolved 4D MRI of abdominal tumors.
METHODS
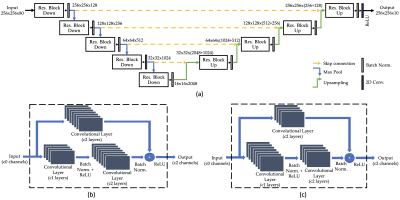
Data acquisition: Seventeen patients with cancer in the abdominal area were scanned on five different 3T scanners (4 Discovery MR750 scanners and 1 Signa Premier scanner, GE Healthcare) using a 3D T1-weighted golden-angle stack-of-stars acquisition (Cartesian kz and radial ky-kx) with the following parameters: TR=3-4ms, TE=1.5-2ms, flip angle = 12°, in-plane resolution = 1.25-1.5mm, slice thickness = 4-5mm, number of radial spokes = 1800, and scan time = 4.3-5 min. XD-GRASP2 reconstruction with 10 motion states was performed on each dataset.MRI-movienet architecture: The proposed MRI-movienet replaces the iterative compressed sensing reconstruction in XD-GRASP. MRI-movienet uses a modified U-net architecture with residual learning blocks to exploit correlations in 5D image space (x, y, z, time, coil) without enforcing data consistency in k-space to remove aliasing artifacts and reconstruct an unaliased motion-resolved 4D image (Figure 1). The network concatenates the coil and time dimensions into a single dynamic dimension (80 combined points = 8 coils x 10 respiratory states) to exploit spatial correlations along both the coil and time dimensions. Skip connections (dashed yellow lines) are used to transfer features at different levels of down-sampling and up-sampling.
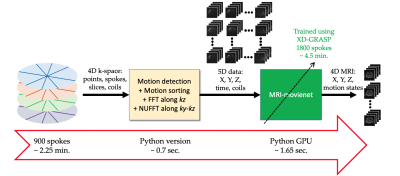
MRI-movienet training: MRI-movienet was trained with an input given by the 5D images corresponding to 900 spokes, which represents a 2-fold acquisition acceleration with respect to XD-GRASP, and target given by XD-GRASP reconstruction with 1800 spokes (Figure 2). 14 datasets were employed for slice-by-slice training (774 slices in total, between 42 and 64 slices per dataset). Network training uses a smooth L1-loss function given by $$$\sum{l_n}$$$, for $$$l_n = 1, \cdots, N$$$, with $$$N$$$ representing the batch size, and, $$$ l_n = \left\{ \begin{array}{ll}\frac{1}{2} \left( x_n - y_n \right)^2, & \text{if } |x_n - y_n| < 1 \\|x_n - y_n| - 0.5, & \text{otherwise},\end{array}\right.$$$, for a given output $$$n_x$$$ and target $$$y_n$$$. This loss function combines the advantages of the L1-loss function, which is less sensitive to the outliers of the L2-loss function.
MRI-movienet testing: Three datasets (patients not included in training) were used to test the performance of MRI-movienet with 900 spokes against the reference XD-GRASP with 1800 spokes. Quantitative image quality evaluation was performed using structural similarity index measure (SSIM)5, peak-signal-to-noise ratio (PSNR), mean squared error (MSE), and normalized root mean squared error (NRMSE) calculated as an average over all the z-slices.
RESULTS
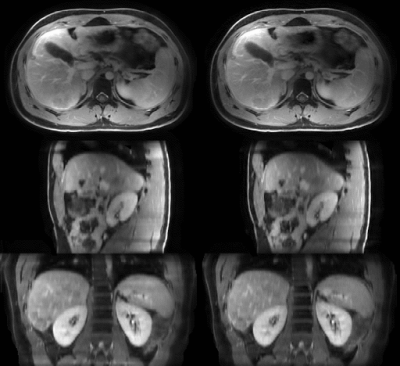
The average MRI-movienet 4D reconstruction time was only 1.65 seconds, which was significantly lower than the iterative XD-GRASP reconstruction time in the order of tens of minutes. The image quality of MRI-movienet is comparable to the one from XD-GRASP, despite the 2-fold acceleration of data acquisition and significantly shorter reconstruction time (Figure 3), which is corroborated by the quantitative evaluation metrics in Figure 4. SSIM was higher than 0.94, and NMRSE was lower than 3%.DISCUSSION
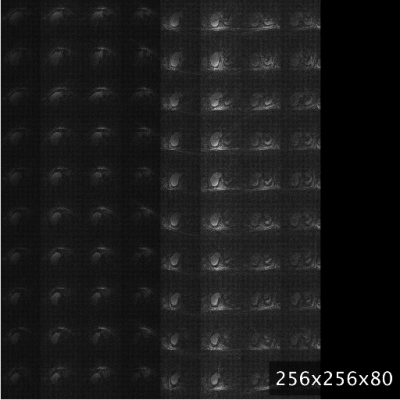
MRI-movienet can remove the use of data consistency without introducing hallucinations by exploiting the fact that motion-resolved images can be represented as a combination of a static background and a dynamic frame-to-frame motion. Even though the dynamic input image is undersampled, there is enough information to estimate the static component (for example, by a simple average), which serves as prior information to the network to avoid hallucinations. Figure 5 shows an example of how MRI-movienet performs reconstruction at different stages of the network. There is a separation of static and dynamic components to remove aliasing artifacts. The presence of the fully-sampled static component, which is passed to decoding layers by the skip connections, helps to avoid hallucinations.CONCLUSION
This work demonstrated the application of a novel deep learning architecture that exploits space-time-coil correlations without k-space data consistency to perform motion-resolved 4D MRI with 2-fold acceleration in data acquisition and significantly faster reconstruction times (about 1.65 seconds) than state-of-the-art XD-GRASP. MRI-movienet would enable 4D MRI with clinically feasible acquisition and reconstruction times, and hence promote routine clinical use.Acknowledgements
The work was supported by NIH Grant R01-CA255661.References
1. Stemkens, Bjorn, Eric S. Paulson, and Rob HN Tijssen. "Nuts and bolts of 4D-MRI for radiotherapy." Physics in Medicine & Biology 63, no. 21 (2018): 21TR01.
2. Feng, Li, Leon Axel, Hersh Chandarana, Kai Tobias Block, Daniel K. Sodickson, and Ricardo Otazo. "XD‐GRASP: golden‐angle radial MRI with reconstruction of extra motion‐state dimensions using compressed sensing." Magnetic resonance in medicine 75, no. 2 (2016): 775-788.
3. Freedman, J. N., Gurney-Champion, O. J., Nill, S., Shiarli, A. M., Bainbridge, H. E., Mandeville, H. C., Koh, D.M., McDonald, F., Kachelrieß, M., Oelfke, U., and Wetscherek, A. (2021). Rapid 4D-MRI reconstruction using a deep radial convolutional neural network: Dracula. Radiotherapy and Oncology, 159, 209-217.
4. Chatterjee, S., Breitkopf, M., Sarasaen, C., Yassin, H., Rose, G., Nürnberger, A., and Speck, O. (2022). ReconResNet: Regularised residual learning for MR image reconstruction of Undersampled Cartesian and Radial data. Computers in Biology and Medicine, 143, 105321.
5. Wang, Zhou, Alan C. Bovik, Hamid R. Sheikh, and Eero P. Simoncelli. "Image quality assessment: from error visibility to structural similarity." IEEE transactions on image processing 13, no. 4 (2004): 600-612.
Figures