0391
Undersampling reconstruction of ferumoxytol-enhanced cardiac cine MRI using a spatiotemporal neural network1Department of Radiological Sciences, University of California Los Angeles, Los Angeles, CA, United States, 2Department of Physics and Biology in Medicine, University of California Los Angeles, Los Angeles, CA, United States, 3MR R&D Collaborations, Siemens Medical Solutions USA, Inc., Los Angeles, CA, United States
Synopsis
Keywords: Machine Learning/Artificial Intelligence, Image Reconstruction
Ferumoxytol can support high quality SGE cardiac cine with blood-myocardial CNR similar to SSFP cine, but free of off-resonance artifact. Much work is ongoing to accelerate the acquisition of multi-slice, breath held cardiac cine. Deep learning-based reconstruction methods can accelerate the image acquisition and reconstruction but needs a large amount of data to train. When compared with compressed sensing and low rank reconstructions, our network showed sharper images and higher consistency with the reference and significantly better quantitative evaluation metrics. We showed that a network trained with non-contrast images could generalize to accelerated ferumoxytol-enhanced cardiac cine MRI with 10x acceleration.INTRODUCTION
Ferumoxytol has increasing applications for cardiovascular MRI in patients with renal impairment and/or congenital heart disease (CHD). Conventionally, multi-slice 2D cine images are collected using ECG-gating during multiple breath-holds, which results in a long acquisition time and possible slice misalignments.1 Parallel imaging (PI) and compressed sensing (CS) reconstruction have been proposed to accelerate image acquisition. However, PI has a limited acceleration rate and CS requires extensive computational resources and a long reconstruction time.2-4 The recent development of deep learning enables more accurate modeling of the image priors and faster reconstruction compared to CS reconstructions.5,6 However, the generalizability of the trained network on contrast-enhanced images has not been investigated. In this work, we aimed to investigate the performance of a neural network trained with non-contrast images on ferumoxytol-enhanced cardiac cine MRI.METHODS
Variable splittingCS reconstruction can be formulated as minimizing the optimization problem:
$$\underset{m}\min\{||Am-y||^2_2 + \lambda R(m)\}$$
where m is the complex-valued MR image, y is the undersampled k-space and A represents coil sensitivity, Fourier transform and undersampling operations. The first term is the data fidelity term, and the second term is the regularization term. Inspired by the fast convergence of the variable splitting method, the optimization problem is transferred to a denoising problem, a data consistency problem and a weighted sum problem.7
Network architecture and training
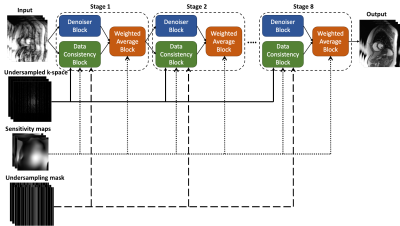
An unrolled spatiotemporal network was developed to integrate the iterative process. As shown in Figure 1, the variable-splitting network has multiple stages, each consisting of a denoiser, a data consistency block and a weighted sum block. The denoiser is learned by a 3D spatiotemporal CNN. The inputs to the network are undersampled 2D cine images, undersampled k-space, sensitivity maps and undersampling masks. The network utilizes mean-squared-error (MSE) loss to minimize the difference between the output and the reference.
The network was trained using 10x undersampled non-contrast cardiac cine MRI and tested using ferumoxytol-enhanced cardiac cine MRI. The network was trained for 26 epochs using an Adam optimizer with momentum β=0.9, mini-batch size of 1, and an initial learning rate of 0.0001. Weights for the network were initiated with random normal distributions with a variance of σ = 0.01 and mean µ=0. The training was completed in Python v3.7 on a graphics processing unit (NVIDIA Tesla V100, 32GB).
Data acquisition and preparation
The study was HIPAA compliant and approved by the institutional review board. To train and evaluate the network, non-contrast 2D SSFP cine images with 30 cardiac phases (2x GRAPPA) were acquired with ECG retrogating in 42 patients on a 1.5T scanner (MAGNETOM Avantofit, Siemens Healthcare, Erlangen, Germany), totally 531 images. The images were divided into 424 training data and 107 validation data. Ferumoxytol-enhanced testing data were acquired using 2D ECG-retrogated GRE sequences from 6 CHD adult patients on the 1.5T scanner (30 phases and 2x GRAPPA) and 6 patients and 5 healthy subjects on a 3T scanner (MAGNETOM Skyra, Siemens Healthcare, Erlangen, Germany, 25 phases and 3x GRAPPA), totally 192 images.
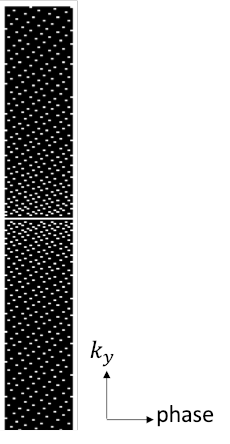
A research application tool written in C++ using the image reconstruction framework of the MRI scanner was used to reconstruct the aliasing-free multi-channel complex images and to extract the sensitivity maps for each image. The images were retrospectively undersampled using variable-density sampling across phase-encoding and cardiac phase dimensions. An example undersampling mask is shown in Figure 2.
Evaluation
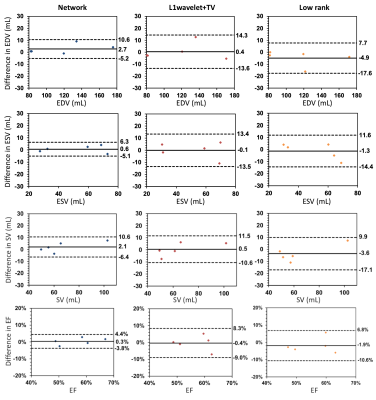
The proposed network was evaluated on retrospectively undersampled images and compared with CS reconstruction with spatial l1-wavelet and temporal total variation (TV) regularizations8 and low rank reconstruction9, both of which were completed using the Bart toolbox10. To evaluate the image quality of different reconstructions, normalized MSE (NMSE), structural similarity index (SSIM) and peak signal-to-noise ratio (PSNR) were computed for all test subjects. Paired t-tests were conducted to determine the statistical significance between the network and other methods. Cardiac function analysis including left ventricular end-diastolic volume (EDV), end-systolic volume (ESV), stroke volume (SV) and ejection fraction (EF) were calculated for 5 CHD patients using cvi42 (Circle Cardiovascular Imaging, Calgary, AB, Canada). The EDV, ESV, SV and EF measured from different methods were compared to those measured from the reference using Bland-Altman analysis with mean difference (MD) and limits of agreement (LoA=MD±1.96×SD).
RESULTS
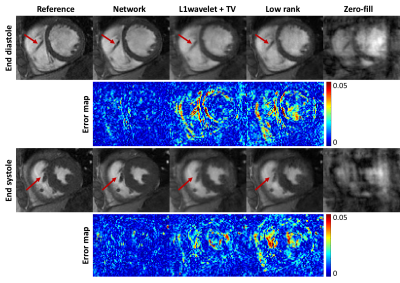
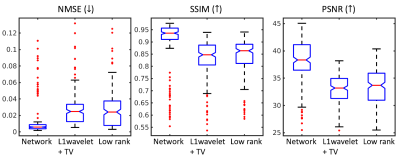
Figure 3 shows the network, CS and low rank reconstructions of a patient with 10x acceleration. All reconstruction methods removed ghosting artifacts and the network showed sharper images and higher consistency compared to CS and low rank reconstructions. NMSE, SSIM and PSNR evaluation results are shown in Figure 4. Paired t-tests show that the proposed network had significantly lower NMSE (p<0.0001), significantly higher SSIM (p<0.0001) and PSNR (p<0.0001) than the other reconstructions. Figure 5 shows the Bland-Altman plots of EDV, ESV, SV and EF from all reconstruction methods. The network produced smaller ranges of LoA compared to the other reconstructions.DISCUSSION AND CONCLUSION
In this study, we developed a spatiotemporal deep neural network to reconstruct ferumoxytol-enhanced accelerated cardiac cine images using the variable splitting method. Results in this study show that even not trained with contrast-enhanced images, the network trained with non-contrast images can be applied to reconstruct artifact-free ferumoxytol-enhanced cardiac cine images using 10x accelerated acquisition. This network may allow more efficient ferumoxytol-enhanced cardiac cine imaging.Acknowledgements
No acknowledgement found.References
[1] Kramer CM, Barkhausen J, Bucciarelli-Ducci C, Flamm SD, Kim RJ, Nagel E. Standardized cardiovascular magnetic resonance imaging (CMR) protocols: 2020 update. Journal of Cardiovascular Magnetic Resonance. 2020 Dec;22(1):1-8.
[2] Lustig M, Donoho D, Pauly JM. Sparse MRI: the application of compressed sensing for rapid MR imaging. Magn Reson Med. 2007;58:1182-1195.
[3] Jung H, Sung K, Nayak KS, Kim EY, Ye JC. k-t FOCUSS: a general compressed sensing framework for high resolution dynamic MRI. Magn Reson Med. 2009 Jan;61(1):103-16. doi: 10.1002/mrm.21757. PMID: 19097216.
[4] Feng L, Srichai MB, Lim RP, Harrison A, King W, Adluru G, Dibella EV, Sodickson DK, Otazo R, Kim D. Highly accelerated real-time cardiac cine MRI using k-t SPARSE-SENSE. Magn Reson Med. 2013 Jul;70(1):64-74. doi: 10.1002/mrm.24440. Epub 2012 Aug 6. PMID: 22887290; PMCID: PMC3504620.
[5] Küstner T, Fuin N, Hammernik K, Bustin A, Qi H, Hajhosseiny R, Masci PG, Neji R, Rueckert D, Botnar RM, Prieto C. CINENet: deep learning-based 3D cardiac CINE MRI reconstruction with multi-coil complex-valued 4D spatio-temporal convolutions. Scientific reports. 2020 Aug 13;10(1):1-3.
[6] Sandino CM, Lai P, Vasanawala SS, Cheng JY. Accelerating cardiac cine MRI using a deep learning‐based ESPIRiT reconstruction. Magnetic Resonance in Medicine. 2021 Jan;85(1):152-67.
[7] Duan J, Schlemper J, Qin C, Ouyang C, Bai W, Biffi C, Bello G, Statton B, O’regan DP, Rueckert D. VS-Net: Variable splitting network for accelerated parallel MRI reconstruction. InInternational conference on medical image computing and computer-assisted intervention 2019 Oct 13 (pp. 713-722). Springer, Cham.
[8] Uecker M, Lai P, Murphy MJ, Virtue P, Elad M, Pauly JM, Vasanawala SS, Lustig M. ESPIRiT--an eigenvalue approach to autocalibrating parallel MRI: where SENSE meets GRAPPA. Magn Reson Med. 2014 Mar;71(3):990-1001. doi: 10.1002/mrm.24751. PMID: 23649942; PMCID: PMC4142121.
[9] Zhang T, Pauly JM, Levesque IR. Accelerating parameter mapping with a locally low rank constraint. Magn Reson Med. 2015 Feb;73(2):655-61. doi: 10.1002/mrm.25161. Epub 2014 Feb 5. PMID: 24500817; PMCID: PMC4122652.
[10] Uecker M, Ong F, Tamir JI, et al. Berkeley advanced reconstruction toolbox. In: Proceedings of 23rd Annual Meeting of the International Society of Magnetic Resonance in Medicine, Toronto, Ontario, Canada, 2015. p. 2486.
Figures