0389
Deep Reconstruction Framework with Self-calibration Mechanisms (DEISM) for Accelerated CEST Imaging1Key Laboratory for Biomedical Engineering of Ministry of Education, Department of Biomedical Engineering, College of Biomedical Engineering & Instrument Science, Zhejiang University, Hangzhou, Zhejiang, China, Hangzhou, China, 2MR Collaboration, Siemens Healthcare Ltd., Shanghai, China, Shanghai, China
Synopsis
Keywords: Machine Learning/Artificial Intelligence, Image Reconstruction
The widespread clinical adoption of chemical exchange saturation transfer (CEST) imaging has been hampered by its prolonged scan time, due to multiple data acquisitions over the varying saturation offset frequencies. In this work, we utilize the artifact suppression algorithm and propose an effective deep reconstruction framework with self-calibration mechanisms (DEISM). The DEISM method was validated on brain tumor patients at 3T. In conjunction with deep-learning multi-coil image reconstruction and data-driven artifact suppression mechanisms, DEISM can provide reliable reconstructions of highly accelerated CEST data, yielding superior performance compared to state-of-the-art methods.Introduction
The widespread clinical adoption of chemical exchange saturation transfer (CEST) imaging has been hampered by its prolonged scan time, due to multiple data acquisitions over the varying saturation frequency offsets. Several strategies have been developed to speed up CEST data acquisition1-5, most of which are based on multi-coil sensitivity encoding6. However, the acceleration factors allowed by these methods are limited by noise amplification and inaccurate estimation of sensitivity maps. Inspired by the artifact suppression (AS) algorithm1,2,5 developed recently, we propose a deep reconstruction framework with self-calibration mechanisms (DEISM) that integrates deep learning and the self-calibration parallel imaging approach for reliable reconstruction of highly accelerated clinical CEST data. The proposed DEISM method was validated on brain tumor patients, with results outperforming state-of-the-art techniques noticeably.Conceptual Framework
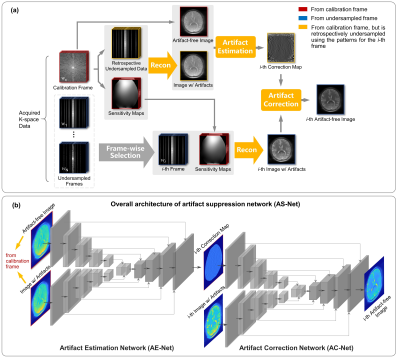
As illustrated in Fig. 1a, a fully-sampled frame is chosen as the calibration one, while the other frames are undersampled. Given that the coil sensitivity is invariable among different saturated frames during CEST imaging, the accurate sensitivity maps calculated from the calibration frame can be shared with the others. However, even though accurate sensitivity maps are used, residual artifacts persist in reconstructed images due to factors like signal leakage caused by the T2 blurring effect5. A two-step artifact suppression algorithm1,2,5 has been derived recently to correct the residual artifacts: (i) Frame-wise Artifact Estimation (AE) using the fully-sampled calibration frame and retrospectively accelerated image from the calibration one, yielding a correction map; (ii) Frame-wise Artifact Correction (AC), in which the residual artifacts in the i-th reconstructed image from the i-th undersampled frame are removed using the corresponding correction map.The existing algorithm simply generates the correction maps by voxel-wise division5. To achieve more robust and effective artifact suppression, a novel deep neural network, dubbed artifact suppression network (AS-Net), is proposed in this work, as illustrated in Fig. 1 (b). The aforementioned AE and AC steps are learned and implemented by two similar convolutional encoder-decoder networks, i.e, AE-Net and AC-Net, both of which are modified from U-Net7.
Methods
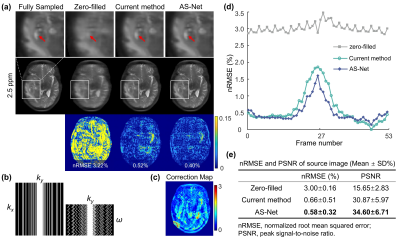
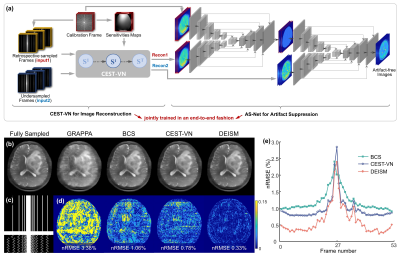
Preliminary Experiment: To independently evaluate the performance of the proposed AS-Net, the process of parallel imaging reconstruction illustrated by “Recon” in Fig. 1a was replaced by inverse Fourier transform after zero-filling. In this experiment, the frame at -3.75 ppm was selected as the calibration one, while the other frames were undersampled with a reduction factor of R=3. Additionally, AS-Net was trained on simulated CEST data8 using a mean squared error (MSE) loss function and tested on real CEST data from brain tumor patients.Proposed Model: The detailed architecture of DEISM is presented in Fig. 3a, which is composed of two main modules: (i) a CEST-VN8,9 that represents the “Recon” node in Fig. 1a and reconstructs images from multi-coil undersampled measurements, with the following formula8,9:
$$\mathbf{S}^{k+1}=\mathbf{S}^k-\gamma^k \mathbf{E}^H\left(\mathbf{E} \mathbf{S}^k-\mathbf{Y}\right)-\sum_{i=1}^{N_v} \mathbf{D}_i^T\left(\varphi_i\left(\mathbf{D}_i \mathbf{S}^k\right)\right),$$
(ii) then an AS-Net estimates and removes the residual artifacts in reconstructed images, and outputs artifact-free images. These two modules are jointly trained in an end-to-end fashion using a CEST-specific loss function8 which jointly measures the error of reconstructed source images and calculated CEST signals:
$$L(\Theta)=\sum_{\left\{\mathbf{Y} \mathbf{s}^*\right\} \in \Omega}\left\|f(\mathbf{Y} \mid \Theta)-\mathbf{S}^*\right\|_2^2+\mu \sum_{\left\{\mathbf{Y}, \mathbf{s}^*\right\} \in \Omega}\left\|\Lambda(f(\mathbf{Y} \mid \Theta))-\Lambda\left(\mathbf{S}^*\right)\right\|_2^2.$$
Imaging Data Acquisition: Human experiments were conducted on a 3T Siemens Prisma MRI system. A single-slice TSE-CEST sequence10 was run with 54 saturation offsets acquired. Plus, a dual-echo GRE sequence was used for B0 field mapping. The trained DEISM was applied to retrospectively undersampled CEST data from 5 glioma patients for validation. And then, the amide proton transfer weighted (APTw) image was calculated by the magnetization transfer ratio asymmetry analysis using source images reconstructed by DEISM.
Results
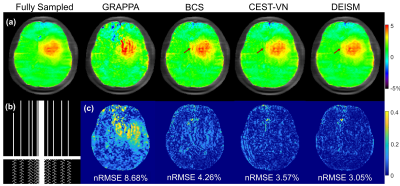
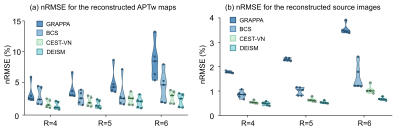
The results of the preliminary experiment are shown in Fig. 2. The artifacts caused by 3-fold undersampling were removed by both AS and AS-Net, without using multi-coil parallel imaging reconstruction. But in comparison with the conventional AS algorithm, the proposed AS-Net provided more details in the image qualitatively, and resulted in lower errors and higher PSNR values quantitatively. These results demonstrated the feasibility and advantage of implementing the AS concept in a neural network manner.The performance of full DEISM with multi-coil reconstruction was then compared with GRAPPA11, BCS12, and CEST-VN8,9 on newly-diagnosed glioma patients. Fig. 3 depicts the fully-sampled and accelerated source image at 3.5 ppm, and associated reconstruction errors at different saturation offsets with acceleration factor R=6. Plus, 5-fold accelerated APTw maps reconstructed by different methods and corresponding error maps are depicted in Fig. 4. In contrast to the other methods, DEISM yielded superior performance in both artifact removal and detail retention, especially for lesion tissues. Fig. 5 illustrates the statistical analyses on five brain tumor patients with varying reduction factors R from 4 to 6, in terms of the nRMSE for the APTw maps and source images reconstructed by different methods, showing DEISM can provide more accurate and reliable reconstruction.
Conclusion
We proposed a novel deep reconstruction framework with self-calibration mechanisms. In conjunction with deep-learning image reconstruction and data-driven artifact suppression mechanisms, DEISM can provide accurate and reliable reconstruction of highly accelerated clinical CEST data, yielding superior performance in terms of both image quality and reconstruction errors compared to the state-of-the-art methods.Acknowledgements
National Natural Science Foundation of China: 81971605. Key R&D Program of Zhejiang Province: 2022C04031. Leading Innovation and Entrepreneurship Team of Zhejiang Province: 2020R01003. This work was supported by the MOE Frontier Science Center for Brain Science & Brain-Machine Integration, Zhejiang University.References
1. Zhang Y, Heo HY, Lee DH, Jiang S, Zhao X, Bottomley PA, Zhou J. Chemical exchange saturation transfer (CEST) imaging with fast variably‐accelerated sensitivity encoding (vSENSE). Magnetic resonance in medicine 2017;77(6):2225-2238.
2. Zhang Y, Heo HY, Jiang S, Zhou J, Bottomley PA. Fast 3D chemical exchange saturation transfer imaging with variably‐accelerated sensitivity encoding (vSENSE). Magnetic resonance in medicine 2019;82(6):2046-2061.
3. Heo HY, Zhang Y, Lee DH, Jiang S, Zhao X, Zhou J. Accelerating chemical exchange saturation transfer (CEST) MRI by combining compressed sensing and sensitivity encoding techniques. Magnetic resonance in medicine 2017;77(2):779-786.
4. She H, Greer JS, Zhang S, Li B, Keupp J, Madhuranthakam AJ, Dimitrov IE, Lenkinski RE, Vinogradov E. Accelerating chemical exchange saturation transfer MRI with parallel blind compressed sensing. Magnetic resonance in medicine 2019;81(1):504-513.
5. Zu T, Sun Y, Wu D, Zhang Y. Joint K-space and Image-space Parallel Imaging (KIPI) for accelerated chemical exchange saturation transfer acquisition. Magnetic Resonance in Medicine 2022;10.1002/mrm.29480.
6. Pruessmann KP, Weiger M, Scheidegger MB, Boesiger P. SENSE: sensitivity encoding for fast MRI. Magnetic Resonance in Medicine: An Official Journal of the International Society for Magnetic Resonance in Medicine 1999;42(5):952-962.
7. Ronneberger O, Fischer P, Brox T. U-Net: Convolutional Networks for Biomedical Image Segmentation. In: Navab N, Hornegger J, Wells WM, Frangi AF, editors; 2015 2015//; Cham. Springer International Publishing. p 234-241.
8. Xu J, Zu T, Hsu Y-C, Sun Y, Wu D, Wang X, Zhang Y. Accelerating Chemical Exchange Saturation Transfer Imaging Using a Model-based Deep Neural Network. arXiv preprint arXiv:220510265 2022.
9. Hammernik K, Klatzer T, Kobler E, Recht MP, Sodickson DK, Pock T, Knoll F. Learning a variational network for reconstruction of accelerated MRI data. Magnetic Resonance in Medicine 2018;79(6):3055-3071.
10. Liu R, Zhang H, Niu W, Lai C, Ding Q, Chen W, Liang S, Zhou J, Wu D, Zhang Y. Improved chemical exchange saturation transfer imaging with real‐time frequency drift correction. Magnetic resonance in medicine 2019;81(5):2915-2923.
11. Griswold MA, Jakob PM, Heidemann RM, Nittka M, Jellus V, Wang JM, Kiefer B, Haase A. Generalized Autocalibrating Partially Parallel Acquisitions (GRAPPA). Magnetic Resonance in Medicine 2002;47(6):1202-1210.
12. Bhave S, Lingala SG, Johnson CP, Magnotta VA, Jacob M. Accelerated whole‐brain multi‐parameter mapping using blind compressed sensing. Magnetic resonance in medicine 2016;75(3):1175-1186.
Figures