0383
Deep Learning based MR Image Reconstruction from Uniformly Undersampled MR Data1Hangzhou Weiying Medical Technology Co., Ltd, Hangzhou, China, 2Guangdong-Hongkong-Macau Institute of CNS Regeneration, Key Laboratory of CNS Regeneration (Ministry of Education), Jinan University, Guangzhou, China
Synopsis
Keywords: Machine Learning/Artificial Intelligence, Image Reconstruction
MR Image reconstruction of uniformly undersampled data often relies on prior information estimated from additional calibration data, leading to compromised acquisition efficiency and flexibility. Here, we propose a joint multi-slice deep learning strategy for MR image reconstruction from uniformly undersampled data with complementary undersampling across adjacent slices. Specifically, we design a slice fusion block to fully exploit the structural and phase similarity in adjacent slices and a slice shift block to further suppress the aliasing artifacts introduced by uniform undersampling. Consequently, the proposed strategy enables accurate MR image reconstruction for both image magnitude and phase without additional calibration information.Introduction
MR Image reconstruction of uniformly undersampled MR data often relies on complete prior information estimated from additional calibration data1,2 (e.g., CSM or ACSs), leading to compromised acquisition efficiency and flexibility. Recently, joint multi-slice MR recontruction3-7 has shown great potential in exploiting the strong correlation among adjacent slices and thus less demanding for calibration data. In this study, we propose a deep learning strategy for MR image reconstruction from multiple consecutive uniformly undersampled data with complementary undersampling across adjacent slices. By utilizing the structural and phase similarity in adjacent slices, which is further enhanced by complementary sampling, the proposed strategy enables accurate MR image reconstruction for both image magnitude and phase without additional calibration information.Method
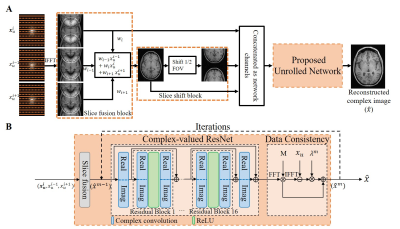
Proposed methodOur preliminary study5-7 explored joint multi-slice PF reconstruction using deep learning. In this study, we propose enhanced joint multi-slice deep learning reconstruction for uniform undersampling with complementary sampling across adjacent slices as depicted in Figure 1, termed EMSDL (SSDL for single-slice deep learning reconstruction). To fully exploit the structural and phase similarity in adjacent slices, a slice fusion block is introduced with learnable parameters to investigate the potential correlation among adjacent slices shown in Figure 1(A). Additionally, a slice shift block is further proposed to suppress aliasing artifacts in regular uniform undersampling, where the periodic aliasing pattern only depends on the acceleration factor. Figure 1(B) presents the proposed iterative or unrolled CNN-based network. Within each iteration, it alternates between reconstructing the MR image through complex image domain residual CNN module (ResNet8) and enforcing the data consistency between the reconstructed image and uniform undersampled data. 3T T1w GRE brain data with 1mm isotropic resolution from Calgary-Campinas Public Brain MR Database9 and 1.5T PDw FSE knee data from fastMRI database10 were used for training and evaluation. The acquisition parameters of brain were TR/TE/TI=6.3/2.6/400 ms and matrix size=226×218×170. The knee datasets were acquired with TR=2200-3000ms, TE=27-34ms, FOV=160×160mm2, matrix size=320×320, and slice thickness/gap=4.5/0mm. All datasets were retrospectively undersampled according to the uniform undersampling schemes. The training was carried out by maximizing the SSIM using Adam optimizer with β1 = 0.99, β2 = 0.999, and initial learning rate = 0.0001. We trained the network with a batch size of 16 for 30 epochs on the Quadro RTX 8000 GPU and Intel Core i9-10900X CPU using PyTorch 1.8.1 packages, which took approximately 13 hours.
Performance Evaluation
We compared the proposed EMSDL method with SSDL method. NRMSE, PSNR, and SSIM were calculated for quantitative evaluation. The reconstructed magnitude and phase images, their corresponding zoomed views, and error maps were examined for a visual comparison.
Results
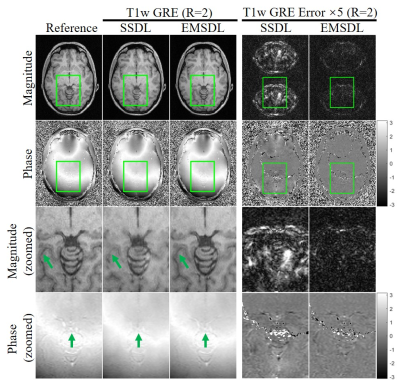
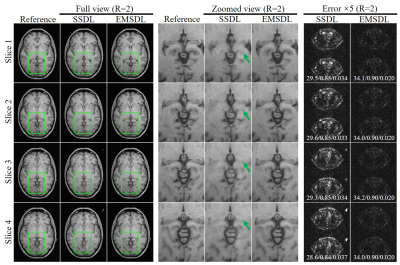
Figure 2 presents the typical results using the proposed SSDL and EMSDL methods for reconstructing the uniformly undersampled data at R = 2 along PE direction (AP). The aliasing artifacts (indicated by green arrows) in SSDL reconstruction were substantially reduced using the EMSDL method by fully utilizing the similarity in structure and phase across adjacent slices. In addition, the EMSDL method could reconstruct images comparable to the fully sampled reference image, as well as the consistently better PSNR, NRMSE, and SSIM values. Significant improvement could be observed in both magnitude and phase images.Figure 3 shows the reconstruction for uniformly undersampled T1w GRE data of the other four adjacent slices at R = 2. The aliasing artifacts (indicated by green arrows) were consistently reduced using the EMSDL method for all slices and significantly outperformed the SSDL methods. The unique features of a certain slice can be well preserved, demonstrating the robustness of the proposed strategy without introducing extra features from other slices.
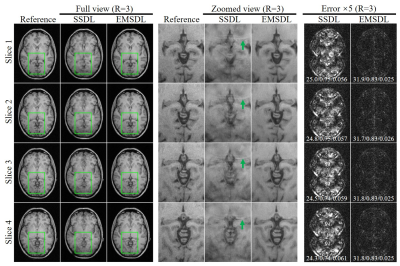
Reconstruction results at a relatively high acceleration factor (R = 3) for single-channel T1w axial GRE of four adjacent slices are shown in Figure 4. Note that the EMSDL method produced a slightly higher error as the acceleration factor increased, while the performance of the SSDL method suffered from a substantial deterioration in reconstructed image quality. This demonstrates the proposed EMSDL enabled a high acceleration factor and was superior to SSDL in suppressing aliasing artifacts.
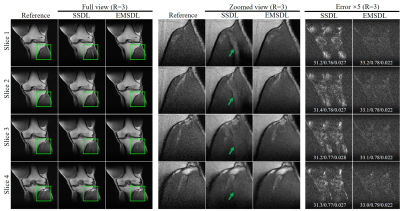
In Figure 5, we applied EMSDL method to PDw FSE knee images at R = 3. The proposed EMSDL method could reconstruct images with less residual aliasing artifacts as well as better PSNR, NRMSE, and SSIM values, indicating the robustness and effectiveness of the proposed EMSDL method.
Discussion and Conclusions
This study demonstrates a joint multi-slice deep learning strategy for accurate MR reconstruction from uniformly undersampled MR data without calibration information. The calibration information is implicitly yet effectively extracted by deep learning with the design of complementary sampling pattern and the slice fusion block, which are expected to fully exploit the similarity in image contents across adjacent slices. Moreover, the periodic aliasing artifacts can be significantly suppressed by the introduction of the slice shift block, which enables the strategy to effectively learn nonlocal correlation. Practically, the complementary sampling pattern for uniform undersampling can be easily implemented by shifting the in-plane phase encoding across adjacent slices or for several acquisitions. Further studies are warranted to verify the effectiveness and robustness of the proposed strategy on the prospectively undersampled data.Acknowledgements
This work was supported in part by Hangzhou Weiying Medical Technology Co., Ltd, and the National Natural Science Foundation of China (82202096).References
[1] Pruessmann KP, Weiger M, Scheidegger MB, Boesiger P. SENSE: sensitivity encoding for fast MRI. Magn Reson Med 1999;42:952-962.
[2] Griswold MA, Jakob PM, Heidemann RM, Nittka M, Jellus V, Wang J, Kiefer B, Haase A. Generalized autocalibrating partially parallel acquisitions (GRAPPA). Magn Reson Med 2002;47(6):1202-1210.
[3] Liu Y, Yi Z, Zhao Y, Chen F, Feng Y, Guo H, Leong ATL, Wu EX. Calibrationless parallel imaging reconstruction for multislice MR data using low-rank tensor completion. Magn Reson Med 2021;85:897-911.
[4] Zhao Y, Yi Z, Liu Y, Chen F, Xiao L, Leong ATL, Wu EX. Calibrationless Multi‐Slice Cartesian MRI via Orthogonally Alternating Phase Encoding Direction and Joint Low‐Rank Tensor Completion. NMR Biomed 2022:e4695.
[5] Xiao L, Liu Y, Yi Z, Zhao Y, Xie L, Cao P, Leong ATL, Wu EX. Partial Fourier reconstruction of complex MR images using complex-valued convolutional neural networks. Magn Reson Med 2022;87(2):999-1014.
[6] Cao P, Xiao L, Liu Y, Zhao Y, Feng Y, Leong ATL, Wu EX. Partial Fourier MRI Reconstruction Using Convolutional Neural Networks. In: Proceedings of the 29th Annual Meeting of ISMRM, Virutual Conference & Exhibition, 2020, p 3627.
[7] Xie L, Liu Y, Xiao L, Cao P, Leong ATL, Wu EX. Enhanced Multi-Slice Partial Fourier MRI Reconstruction Using Residual Network. In: Proceedings of the 30th Annual Meeting of ISMRM, Virutual Conference & Exhibition, 2021, p 1976.
[8] Kaiming H, Xiangyu Z, Shaoqing R, Jian S. Deep Residual Learning for Image Recognition. In: IEEE international conference on computer vision and pattern recongnition, 2016, p 770-778.
[9] Souza R, Lucena O, Garrafa J, Gobbi D, Saluzzi M, Appenzeller S, Rittner L, Frayne R, Lotufo R. An open, multi-vendor, multi-field-strength brain MR dataset and analysis of publicly available skull stripping methods agreement. NeuroImage 2018;170:482-494.
[10] Knoll F, Zbontar J, Sriram A, Muckley MJ, Bruno M, Defazio A, Parente M, Geras KJ, Katsnelson J, Chandarana H, Zhang Z, Drozdzalv M, Romero A, Rabbat M, Vincent P, Pinkerton J, Wang D, Yakubova N, Owens E, Zitnick CL, Recht MP, Sodickson DK, Lui YW. fastMRI: A Publicly Available Raw k-Space and DICOM Dataset of Knee Images for Accelerated MR Image Reconstruction Using Machine Learning. Radiol Artif Intell 2020;2:e190007.
Figures