0363
A 3D Fast Multiphase DENSE MRE Sequence with Stack-of-Star Radial Sampling, Interleaved Multi-Slab Acquisition and Hadamard Encoding
Yu Chen1, Runke Wang1, Ruokun Li2, Fuhua Yan2, and Yuan Feng1,2,3
1Shanghai JiaoTong University, School of Biomedical Engineering, Shanghai, China, 2Ruijin Hospital, Shanghai Jiaotong University School of Medicine, Shanghai, China, 3Institute of Medical Robotics, Shanghai Jiao Tong University, Shanghai, China
1Shanghai JiaoTong University, School of Biomedical Engineering, Shanghai, China, 2Ruijin Hospital, Shanghai Jiaotong University School of Medicine, Shanghai, China, 3Institute of Medical Robotics, Shanghai Jiao Tong University, Shanghai, China
Synopsis
Keywords: Elastography, Brain
Conventional gradient echo (GRE) based Magnetic Resonance Elastography (MRE) suffers from long TE and scan time at low mechanical excitation (<25Hz). Displacement Encoding with a Stimulated Echo (DENSE) can record displacement with short TR, useful for low-frequency excitation. Here, 3D excitation was used to compensate for potential signal loss. A multiphase acquisition scheme with stack-of-star radial sampling was also used along with interleaved multi-slab acquisition and Hadamard encoding for MRE acceleration. Phantom experiments were conducted for validation. 32 slices brain MRE scan at 20Hz can be achieved in 11min30s, 5 folds faster than conventional GRE-based MRE.Summary of Main Findings
A 3D GRE-based fast Multiphase DENSE MRE sequence with stack-of-star radial sampling, interleaved multi-slab acquisition and Hadamard encoding was proposed for MRE acceleration. Compared with conventional GRE-based MRE, the proposed sequence can achieve 5-fold acceleration.Introduction
Magnetic resonance elastography (MRE) is a useful method to detect the biomechanical properties of soft tissues noninvasively1, 2. Conventional gradient-echo (GRE) based MRE sequence has to be synchronized with mechanical excitation3, elongating the scanning time when the excitation frequency is low (<25Hz). Displacement encoding with stimulated echo (DENSE) can record displacement with short TR. Therefore, DENSE is ideal for low-frequency encoding than conventional GRE-MRE at low mechanical excitation scenarios. It has been shown that multiphase DENSE MRE with radial sampling has the advantage of acceleration and artifact suppression4. In this study, we proposed a 3D GRE-based fast multiphase DENSE MRE sequence with stack-of-star radial sampling, interleaved multi-slab acquisition5, 6, and Hadamard encoding7 for acceleration of MRE. Phantom experiments were conducted for validation. A human brain MRE scan was also performed to demonstrate its potential for MRE acceleration.Methods
The DENSE method can be divided into two parts: Encode and Decode. Spatial modulation of magnetization block was used for encoding the initial place. In the imaging block, a decode gradient was used to generate a DENSE echo.$$M_{x y}^{D E N S E}=\frac{1}{2} M_{0} \sin \theta \cdot e^{-\frac{T M}{T_{1}}} \cdot e^{i \mathrm{~K} \Delta \mathrm{r}}$$ where $$$M_0$$$ is the steady state longitudinal magnetization, $$$\theta$$$ is the flip angle, $$$TM$$$ is the mixing time, $$$K$$$ is the area under the motion encoding gradient and $$$\Delta r$$$ represents the displacement.
The proposed sequence timing diagram is shown in Figure 1. Motion encoding block (MEB) encoded the initial position and motion decoding blocks (MDB) decoded and generated the DENSE echo. In MEB, the encoding gradients were set up in three encoding directions simultaneously for Hadamard encoding. 1 MEB was followed by N times MDB (N = slice phase encoding (SPE) number * slice oversample rate / 2). One MEB plus N MDB is called an imaging section, which filled the SPE-Spokes k-space a half from the center to the edge. In one MDB, 8 times in plane golden-angle radial sampling acquisition was performed for imaging 4 motion phases for the 2 imaging slabs respectively. A broad-band minimum-phase RF pulse is used for 3D excitation. The decoding gradient was also set up ib three encoding directions simultaneously. The Hadamard DENSE signal was composited of three direction displacements. With 4 times Hadamard encoding, the origin displacement can be decoded using the least square method.
$$M_{x y}^{DENSE}=\frac{1}{2} M_{0} \sin \theta \cdot e^{-\frac{T M}{T_{1}}} \cdot e^{i\left(\mathrm{~K}_{\mathrm{x}} \Delta \mathrm{x}+\mathrm{K}_{\mathrm{y}} \Delta \mathrm{y}+\mathrm{K}_{\mathrm{z}} \Delta \mathrm{z}\right)}$$ $$\left[\begin{array}{l}\Phi_{1} \\\Phi_{2} \\\Phi_{3} \\\Phi_{4}\end{array}\right]=\left[\begin{array}{llll}+1 & +1 & +1 & +1 \\-1 & +1 & +1 & +1 \\+1 & -1 & +1 & +1 \\+1 & +1 & -1 & +1\end{array}\right]\left[\begin{array}{c}K_{x} \Delta x \\K_{y} \Delta y \\K_{z} \Delta z \\\Phi_{0}\end{array}\right]=E\left[\begin{array}{c}K_{x} \Delta x \\K_{y} \Delta y \\K_{z} \Delta z \\\Phi_{0}\end{array}\right]$$ $$\left[\begin{array}{c}\Delta x \\\Delta y \\\Delta z \\\sim\end{array}\right]=\operatorname{inv}(E)\left[\begin{array}{l}\Phi_{1} \\\Phi_{2} \\\Phi_{3} \\\Phi_{4}\end{array}\right] \cdot *\left[\begin{array}{c}1 / K_{x} \\1 / K_{y} \\1 / K_{z} \\\sim\end{array}\right]$$ where $$$K_x, K_y, K_z$$$ are the areas of the encoding gradients on three axes respectively and $$$\Delta x, \Delta y, \Delta z$$$ represent the displacements of three directions. $$$\Phi_i$$$ is the $$$i^{th}$$$ time composite phase with Hadamard encoding.
The proposed sequence was implemented and tested on a 3T scanner (uMR 790, United Imaging Healthcare, Shanghai, China). A custom-built electromagnetic actuator was used to generate the vibration. The imaging parameters were TR=12.5ms, TE=4.2ms, matrix size=80*80, slice=32, voxel size=3*3*3 mm, spokes /slab=125, BW=500Hz/Pixel, $$$G_{MEG}$$$=35mT/m, $$$G_{dura}$$$=0.85ms. A phantom with two stiff inclusions,(I) was stiffer than (II) (Figure 2), was constructed for sequence validation at 60Hz8. A healthy volunteer was recruited for testing the proposed sequence at 20Hz.
Results
Phantom experiments result showed that the displacement field recorded using the proposed was similar to that from GRE-MRE sequence. (Figure 2) Moreover, the two stiff inclusions can also be detected and distinguished from each other in the inversion shear modulus map (Figure 3). The human brain experiment showed the proposed sequence can clearly record the displacement and the potential in GRE-based MRE acceleration with low mechanical excitation. (Figure 4)Conclusion
We proposed a 3D GRE-based fast multiphase DENSE MRE sequence with stack-of-star radial sampling, interleaved multi-slab acquisition and Hadamard encoding for MRE acceleration. The phantom experiment result showed wave images acquired were similar to that of GRE-MRE. The human brain scan showed its potential for acceleration with low-frequency mechanical excitation.Acknowledgements
Funding support from the National Natural Science Foundation of China (grant 32271359) and the Natural Science Foundation of Shanghai (grant 22ZR1429600) is acknowledged.References
[1] S. K. Venkatesh, "Clinical Applications of Liver Magnetic Resonance Elastography: Chronic Liver Disease," in Magnetic Resonance Elastography, 2014, ch. Chapter 4, pp. 39-60.[2] L. V. Hiscox et al., "Magnetic resonance elastography (MRE) of the human brain: technique, findings and clinical applications," Phys Med Biol, vol. 61, no. 24, pp. R401-R437, Dec 21 2016, doi: 10.1088/0031-9155/61/24/R401.
[3] J. Strasser, M. T. Haindl, R. Stollberger, F. Fazekas, and S. Ropele, "Magnetic resonance elastography of the human brain using a multiphase DENSE acquisition," Magn Reson Med, vol. 81, no. 6, pp. 3578-3587, Jun 2019, doi: 10.1002/mrm.27672.
[4] R. K. Wang et al., "Fast magnetic resonance elastography with multiphase radial encoding and harmonic motion sparsity based reconstruction," (in English), Phys. Med. Biol., Article vol. 67, no. 2, p. 13, Jan 2022, Art no. 025007, doi: 10.1088/1361-6560/ac4a42.
[5] P. Garteiser et al., "Rapid acquisition of multifrequency, multislice and multidirectional MR elastography data with a fractionally encoded gradient echo sequence," NMR Biomed, vol. 26, no. 10, pp. 1326-35, Oct 2013, doi: 10.1002/nbm.2958.
[6] C. Guenthner, S. Sethi, M. Troelstra, A. S. Dokumaci, R. Sinkus, and S. Kozerke, "Ristretto MRE: A generalized multi-shot GRE-MRE sequence," NMR Biomed, vol. 32, no. 5, p. e4049, May 2019, doi: 10.1002/nbm.4049.
[7] C. L. Dumoulin, S. P. Souza, R. D. Darrow, N. J. Pelc, W. J. Adams, and S. A. Ash, "Simultaneous acquisition of phase-contrast angiograms and stationary-tissue images with Hadamard encoding of flow-induced phase shifts," (in English), Journal of magnetic resonance imaging : JMRI, vol. 1, no. 4, pp. 399-404, 1991 1991, doi: 10.1002/jmri.1880010403.
[8] Y. Feng et al., "Viscoelastic Characterization of Soft Tissue-Mimicking Gelatin Phantoms using Indentation and Magnetic Resonance Elastography," (in English), J. Vis. Exp., Article no. 183, p. 12, May 2022, Art no. e63770, doi: 10.3791/63770.
Figures
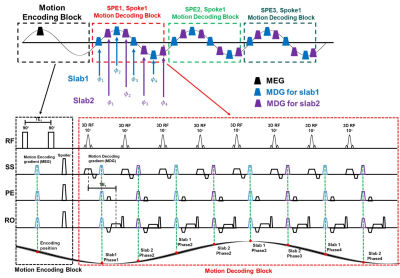
Figure
1
Sequence diagram for
the proposed 3D GRE-based fast Multiphase DENSE MRE sequence. 1 motion encoding
block (MEB) was followed by N times motion decoding block (MDB) (N = slice
phase encoding (SPE) number * slice oversample rate / 2). One MEB plus N MDB is
called an imaging section, which filled the SPE-Spokes k-space a half from the
center to the edge. In one MDB, 8 times in plane golden-angle radial sampling
acquisition was performed for imaging 4 motion phases for the 2 imaging slabs
respectively.
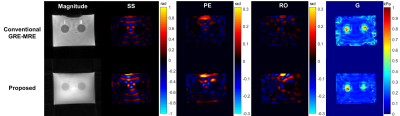
Figure
2
Phantom experiment validation
at 60Hz mechanical excitation. Similar displacement field was recorded by
conventional GRE-MRE and the proposed sequence. The two stiff inclusions can be
detected and distinguished from each other in the inversion shear modulus map.

Figure
3
Statistical
comparison of phantom inversion shear modulus between conventional
GRE-MRE and proposed sequence. The proposed new sequence can better distinguish
inclusions and the background.
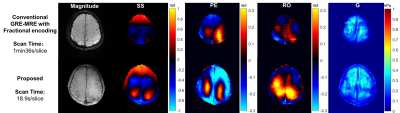
Figure
4
20 Hz Human brain MRE
scan results. Mechanical wave was clearly recorded by the proposed sequence.
The scan time of the proposed sequence showed the potential in GRE-based MRE
acceleration with low mechanical excitation.
DOI: https://doi.org/10.58530/2023/0363