0355
Automatic segmentation of the interscapular BAT in rats by the dynamic magnetic resonance fat fraction images1Shenzhen institutes of advanced technology, Chinese Academy of Sciences, Shenzhen, China, 2Radiology Department, Bethune First Hospital of Jilin University, Changchun, China
Synopsis
Keywords: Segmentation, Fat, brown adipose tissue
The noninvasive assessment of BAT volume is fundamental for characterizing the longitudinal effects in rodents. In this work, dynamic fat fraction images of 34 rats fed under different conditions were acquired before and after noradrenaline injection for 2.5 hours. The iBAT regions were recognized and labelled automatically by identifying the regions with significant changes in FF images. Then a deep learning network was built up by training the FF images and the automatically identified mask images in all rats. The dice similarity coefficient, precision rate and recall rate of the network were found to be 0.897±0.061, 0.901±0.068 and 0.889±0.086, respectively.
Introduction
As an energy-consuming organ, brown adipose tissue (BAT) has been found great potentials in regulating the energy metabolism and fighting against the obesity threaten to health1. The noninvasive assessment of the BAT recruitment as well as its metabolic activity are fundamental for characterizing the longitudinal effects in rodents. Several studies have proposed various methods to identify BAT deposit through the multi-parametric MR images in rodent studies2.Nevertheless, BAT is a highly heterogeneous organ. The physiological features of BAT in rodents depends firmly on several factors, such as age, diet, feeding temperature, and stimulation condition. As a result, the imaging parameters, e.g. fat fraction (FF), of BAT depot in these rats might show very little difference to the surrounding subcutaneous WAT. This challenge brings in great difficulties in the developing segmentation tools for BAT deposit identification due to the ambiguous boundaries in the MR images.
To overcome this challenges, a novel automatic iBAT recognition and labeling method was developed by identifying those regions with significant changes found in the quantitative MR images in response to the β3-agonist stimulation. Also, MR images of various rat models raised in different conditions were collected to simulate the heterogeneity of iBAT. Finally, a neural network was built up by training the quantitative MR images and the automatically identified mask images in all rats.
Methods
The animal studies introduced below were approved by our institute. 34 Sprague Dawley (SD) rats were fed in different conditions. Eighteen rats (dataset1) aged 7 weeks were randomly divided into three groups (6 rats/group) and housed at 30°C, 23°C and 15°C ambient temperatures for 2 weeks in a temperature-controlled incubator (LP-80LED-6CTAR, NK systems, Tokyo, Japan). The rats were intraperitoneally injected with 1mg/kg dose of NE during the MRI scan. Another 16 rats (dataset2) with averaged weight 640g ±28.5g were randomly divided into three groups with different dose of NE injection: 5 rats of 2mg/kg NE, 6 rats of 1mg/kg NE and 5 rats of 0.5mg/kg NE. For all of these 34 rats, MR images were acquired at multiple time points with one scan before NE injection and 30 scans after NE injection in a total of 155 minutes for each rat.MR images were collected using a 3.0T clinical MRI scanner (uMR790, Shanghai United Imaging Healthcare, Shanghai, China). Eight-echo gradient echo imaging sequence was used for image acquisition in the transverse orientation with TE1/ΔTE = 2.99/1.79ms, pixel resolution= 0.47 × 0.47mm2, and slice thickness 2.1mm. The imaging were repeated for 31 times and the scan time was 5 minutes for each acquisition. The complex multi-echo images were processed by our previous proposed fat and water separation algorithm to generate fat fraction images3.
In the present study, the iBAT region was recognized automatically by comparing the FF images of different time points before and after NE injection with significant FF changes, while other regions were labelled as background, including WAT, muscle, bone marrow and other tissues (Figure 1). Then, a deep learning network was trained based on the above automatic iBAT recognition results. The 34 rats were divided randomly into training set, validation set and testing set. The numbers of images in the training set, validation set and testing set are 3968 (22 rats), 1302 (6 rats) and 1240 (6 rats), respectively. For network training, we employed a U-net with EfficientNet as backbone for iBAT segmentation model and the weighted cross-entropy was chosen as loss function in our training4,5. Three evaluation metrics were chosen to evaluate the training network: dice similarity coefficient (DSC), precision rate (PR) and recall rate (RR).
Results
Table 1 shows the mean DSC, PR and RR of the network in the testing set. Figure 2 shows the automatic segmentation results of four representative rats from testing datasets in comparison with the automatically recognized labels. The main differences between the label images and the auto-segmented images exist in the boundary of the iBAT area, which may be attributed to the partial volume effect (PVE) between iBAT and the surrounding white adipose tissue, muscle or other non-adipose tissues.Discussion and conclusions
In this study, an automatic identification and segmentation method of iBAT in rats using FF images was proposed based on deep learning. Compared to previous works, our method is featured in two aspects. First, the iBAT labeling was automatically implemented by identifying the regions where there was significant fat fraction change in response to NE administration. Second, the trained images were collected from a number of rats under various feeding conditions and at multiple time points after activation to simulate the heterogeneity of iBAT. The main difference between labelled mask images and auto-segmented mask images exists in the boundary of iBAT and the surrounding tissues, which could be explained by the partial volume effect (PVE) with pixel resolution of 0.47mm. This might be improved by using the higher-resolution images at ultra-high field MR scanner. As a conclusion, an automatic segmentation method for iBAT region in rats using auto-labelling procedure combined with deep learning network was proposed.Acknowledgements
This research was supported by the National natural Science Foundation of China: 61901462 and the Natural Science Foundation of Guangdong Province: 2022A1515010162.References
[1] Wang W, Seale P. Control of brown and beige fat development[J]. Nature reviews Molecular cell biology, 2016, 17(11): 691-702.
[2] Bhanu Prakash K N, Verma S K, Yaligar J, et al. Segmentation and characterization of interscapular brown adipose tissue in rats by multi-parametric magnetic resonance imaging[J]. Magnetic Resonance Materials in Physics, Biology and Medicine, 2016, 29(2): 277-286.
[3] Cheng C, Zou C, Liang C, et al. Fat‐water separation using a region‐growing algorithm with self‐feeding phasor estimation[J]. Magnetic resonance in medicine, 2017, 77(6): 2390-2401.
[4] Tan M, Le Q. Efficientnet: Rethinking model scaling for convolutional neural networks[C]//International conference on machine learning. PMLR, 2019: 6105-6114.
[5] Aurelio Y S, de Almeida G M, de Castro C L, et al. Learning from imbalanced data sets with weighted cross-entropy function[J]. Neural processing letters, 2019, 50(2): 1937-1949.
Figures
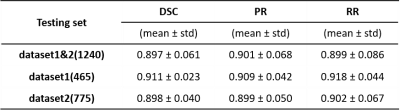
Table 1.Performance evaluation of the trained network
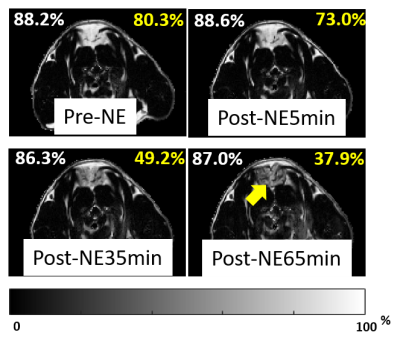
Figure 1. Dynamic FF images before and after the administration of β3 agonist in one rat. A rat aged 22 weeks housed at thermoneutral environment underwent a dosage of 2mg/kg NE. The mean fat fraction of WAT and iBAT area shown in upper left and upper right corners of each image in white and yellow respectively. The FF was expected to decrease significantly after the acute activation of iBAT, whereas no obvious change in FF value of the surrounding subcutaneous WAT. The yellow arrow indicates the iBAT regions.
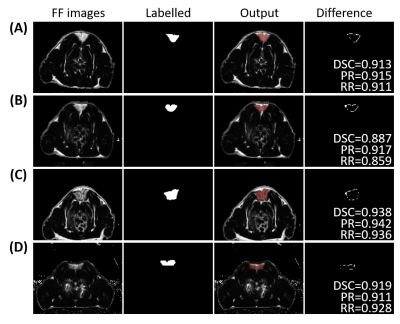
Figure 2. Comparison of the automatic segmentation results and the labels. The first column, FF images of iBAT; the second column, the corresponding label images generated by identifying whether there is significant change before and after NE injection; the third column, auto-segmented iBAT mask images, overlaid on FF images; and the fourth column, the difference between the auto-segmented results and the labels with the quantitative DRC, PR and RR.