0340
Joint cardiac $$$T_1$$$ mapping and cardiac cine using a deep manifold framework1University of Texas Southwestern Medical Center, Dallas, TX, United States, 2University of Iowa, Iowa City, IA, United States, 3University of Wisconsin–Madison, Madison, WI, United States
Synopsis
Keywords: Heart, Machine Learning/Artificial Intelligence, Reconstruction
The main focus of this work is to introduce a deep generative model for simultaneous free-breathing cardiac $$$T_1$$$ mapping and CINE MRI. The data is acquired by a gradient echo inversion recovery sequence with intermittent delays for magnetization recovery. The joint reconstruction of the image time-series is performed using a patient-specific deep manifold reconstruction algorithm which learns a CNN generative model and its latent vectors from the measured k-t space data in an unsupervised fashion. Following learning, the model can be used to generate synthetic images at specific motion and contrast states.Background
Clinical myocardial $$$T_1$$$ mapping [1,2] and functional imaging using SSFP sequences involve breath-held, which is challenging for many patient groups. In addition, the acquisition of these datasets often involves a long scan time, which increases patient inconvenience and increased medical costs. Recently, some researchers have proposed to combine the two acquisitions using ideas similar to MR fingerprinting. For instance, MR multi-tasking [3] relies on a low-rank tensor model for the multi-dimensional signal to relax the breath-holding requirement. The need for dedicated navigators in this approach makes it challenging to adapt to general sequences. In addition, the use of continuous acquisition may make it difficult to decouple the effects of flip angle and $$$T_1$$$ recovery, which may affect the accuracy of the estimated $$$T_1$$$ maps. To overcome this issue, Zhou et. al. [4] introduced a dual flip angle approach, where the acquisition consists of two blocks of two different flip angles. They perform a low-rank and sparse joint recovery of cardiac phase and $$$T_1$$$ dynamics that is conceptually similar to [5].Methods
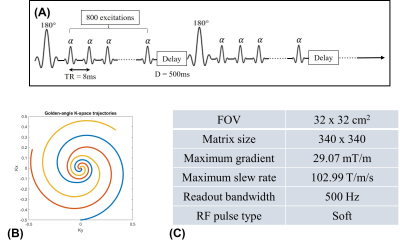
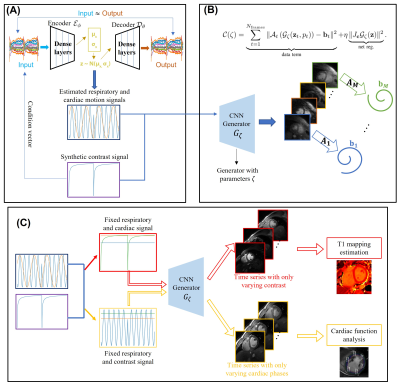
This work introduces a deep manifold framework for the joint recovery of inversion recovery prepared free-breathing and ungated cardiac MRI. The free-breathing and ungated data acquired for joint cardiac $$$T_1$$$ mapping and cardiac cine are based on an inversion recovery sequence. The details of the sequence are depicted in Fig. 1.We model the image frames in the time series as a non-linear function of three variables: cardiac and respiratory phases and inversion time. The non-linear function is realized using a convolutional neural network (CNN) generator, while the CNN parameters and the phase information are estimated from the measured k-t space data. We use a dense conditional auto-encoder to estimate the cardiac and respiratory phases from the central multi-channel k-space samples acquired at each frame. The latent vectors of the auto-encoder are constrained to be bandlimited functions with appropriate frequency bands, which enables the disentanglement of the latent vectors into cardiac and respiratory phases, even when the data is acquired with intermittent inversion pulses. Once the phases are estimated, we pose the image recovery as the learning of the parameters of the CNN generator from the measured k-t space data. The learned CNN generator is used to generate synthetic data on demand, by feeding it with appropriate latent vectors. The framework enables the generation of synthetic breath-held CINE movies with different inversion contrasts as well as the estimation of the $$$T_1$$$ maps with specific phases.
After the data was acquired, we used a two-step data processing strategy to jointly obtain the cardiac $$$T_1$$$ mapping and cardiac cine. In the first step, we try to estimate the cardiac and respiratory motions from the central k-t space data using a conditional variational auto-encoder (VAE). Since the timing of the inversion pulses is known apriori, we feed the inversion timing signal as a conditional vector to the network. Together with the known inversion timing signal, the three latent vectors (cardiac and respiratory motion signals from the VAE and the inversion timing signal) are used in the second step, where the reconstruction of the free-breathing and ungated cardiac MR images happens. For the reconstruction, we model the image frames in the time series as the output of a CNN generator, and the input of the CNN generator is the three latent vectors as discussed above. The idea of the method is illustrated in Fig. 2.
Results
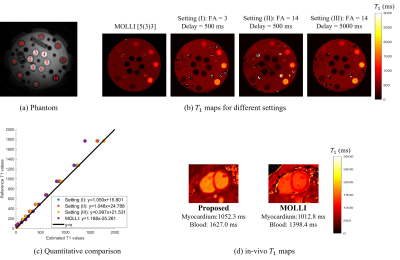
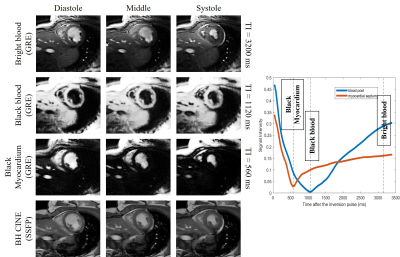
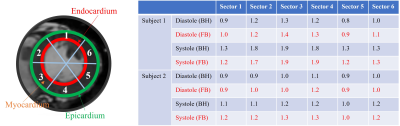
We first show the validation of the $$$T_1$$$ mapping using the proposed scheme. Phantom studies were performed in a commercially-available (Caliber MR, Boulder, CO, USA) phantom shown in Fig. 3 (a). We used the proposed inversion recovery sequence with three different settings: (I), flip angle $$$\alpha = 3^\circ$$$ with a delay of 500 ms. (II), flip angle $$$\alpha = 14^\circ$$$ with a delay of 500 ms. (III), we flip angle $$$\alpha = 14^\circ$$$ with delay of 5000 ms. The conventional 2D PPG-triggered MOLLI data was also acquired for comparison. The results of the phantom study are shown in Fig. 3. We note that the $$$T_1$$$ values roughly match the MOLLI measures in the myocardium, while the estimated $$$T_1$$$ values in the blood-pool are higher than MOLLI. This systematic bias can be explained by the phantom measurements, which show that MOLLI underestimates the high $$$T_1$$$ values.Synthetic breath-held CINE images with different contrast (i.e., at different inversion times) can also be generated from the deep manifold reconstruction algorithm. Specifically, after the training of the generator for each subject, we can fix the respiratory signal and also choose a specific contrast for breath-hold CINE generation with the chosen contrast. The results of synthetic breath-held CINE image generation are shown in Fig. 4. The left ventricle wall analysis based on the generated synthetic breath-held CINE image compared to the breath-held bSSFP images is shown in Fig. 5.
Conclusion
In this study, we proposed a manifold-based recovery scheme for the joint recovery of inversion recovery-prepared free-breathing and ungated cardiac MRI. The framework enables the generation of CINE images with different contrast as well as the estimation of the T1 maps with specific phases.Acknowledgements
No acknowledgement found.References
[1] D. R. Messroghli et. al., “Modified look-locker inversion recovery (molli) for high-resolution t1 mapping of the heart,” MRM, 2004.
[2] K. Chow et. al., “Saturation recovery single-shot acquisition (sasha) for myocardial t1 mapping,” MRM, 2014.
[3] A. G. Christodoulou et. al., “Magnetic resonance multitasking for motion-resolved quantitative cardiovascular imaging,” Nat. Biomed. Eng, 2018.
[4] R. Zhou et. al., “Dual-excitation flip-angle simultaneous cine and t1 mapping using spiral acquisition with respiratory and cardiac self-gating,” MRM, 2021.
[5] J. I. Hamilton et. al., “Mr fingerprinting for rapid quantification of myocardial t1, t2, and proton spin density,” MRM, 2017.
Figures