0339
Highly-Accelerated High-Frame-Rate Cine for Exercise Cardiac MRI
Manuel Morales1, Manuel A Morales1, Siyeop Yoon1, Jennifer Rodriguez1, Warren J Manning1, and Reza Nezafat1
1BIDMC, Boston, MA, United States
1BIDMC, Boston, MA, United States
Synopsis
Keywords: Heart, Machine Learning/Artificial Intelligence
Combined cardiac MRI with exercise (Ex-CMR) is a stress test with promising applications. However, standard ECG-segmented cine imaging during exercise is challenging. Free-breathing ECG-free real time cine can be achieved with compressed sensing. Yet tradeoff remains between temporal and spatial temporal resolution. Thus, we sought to develop a highly accelerated high-frame-rate cine for Ex-CMR by accelerating spatial resolution using Resolution Enhancement Generative Adversarial Inline Network (REGAIN), followed by synthesizing new frames using Deformation ENcoding Transformer (DENT). REGAIN enabled 14-fold scan acceleration, DENT enabled 2-fold improvement in temporal resolution. We achieved spatiotemporal resolution of 1.9 × 1.9 mm2 and 16 ms.Background
Exercise cine MRI is a stress test with promising applications in cardiovascular disease. However, standard ECG-segmented cine imaging during exercise is challenging. Free-breathing ECG-free real time cine can be achieved by combining highly accelerated imaging with compressed sensing (CS). Yet a tradeoff remains between temporal and spatial temporal resolution in cine. In this study, we sought to develop a highly accelerated high-frame-rate cine for exercise cardiac MRI by accelerating spatial resolution using Resolution Enhancement Generative Adversarial Inline Network (REGAIN), followed by synthesizing new cardiac frames using Deformation ENcoding Transformer (DENT). The REGAIN enables 14-fold scan acceleration and DENT enabled 2-fold improvement in temporal resolution, allowing a nominal spatial resolution of 1.9 × 1.9 mm2 and temporal resolution of 16 ms.Methods
Transformer Network for high-frame-rate cine DENT worked in the image domain and generated forward-backwards mappings describing the underlying deformation of multiple cardiac phases. Such deformation was used to synthesize images with high frame rate. Input was W × H cine images (T = 4) collected with temporal resolution ∆t at t - ∆t, t, t + ∆t and t + 2∆t. Output was an interpolated image at time t + 0.5∆t (Fig. 1a). Embedding layer extracted F = 32 features per pixel from inputs. Downsampling step reduced image dimensionality by half while doubling the feature dimensionality. Inputs to transformer layers were split into windows. Multiscale attention was used to learn a spatiotemporal correspondence within each window. For spatial attention, the input was split into N = W•H•T / M2 windows of size M2 × F along the spatial dimension, where M = 8. For temporal attention, the input was split into N = W•H windows of size T × F. Decoder layers upsampled the dimensionality W × H of the encoder output by 2 while reducing the number of features F by half. These were passed to the motion synthesizer block. Additional convolutional layers in the block upsampled the input while generating deformation and scaling components. The new cardiac frame was synthesized from the 4 input frames using these components for sampling and interpolation. The outputs from each resolution scale were combined by upsampling and merging sequentially. Multi-center (centers = 3), multi-vendor (GE, Philips, Siemens), multi-field strength (1.5T, 3T) scans from 3178 patients (2139 male, 54 ± 16 years) undergoing clinical MRI for different cardiac indications were used for training. Cine images were collected using a breath-hold ECG-gated segmented SSFP at 1.5T (n = 1831) and 3T (n = 1347) in short axis and 2-, 3- and 4-chamber. A sample was as a cine slice with T ≥ 7 frames; an epoch one optimization loop across all training samples. First, a center-frame was randomly selected and used as the ground-truth. Second, 4 frames adjacent to were selected as inputs: either or , which had a 2∆t and 4∆t temporal resolution, accordingly (Fig. 1b). Each cine frame was normalized by min-max prior to processing. Ground-truth and input images were randomly cropped to 256 × 256. Training used a batch size = 10 for 100 epochs (~200 hours) using an AdaMax optimizer. Learning rate was initially 2 × 10-2 and gradually decayed to 1 × 10-6. Generative Adversarial Network for enhanced spatial resolution REGAIN enabled additional acceleration (i.e., beyond CS-enabled) by prescribing 2-fold reduced spatial resolution along phase-encode (PE). The low-resolution image was then reconstructed using CS with zero-padding to create an image with full resolution, albeit with significant blurring. Subsequently, REGAIN enhances spatial resolution as analogous to collecting a full-resolution image. The generator of REGAIN enhances the image resolution, and the discriminator classifies the generator's output and high-resolution image for the adversarial training (Fig 2). To allow inline reconstruction, REGAIN was seamlessly integrated with the scanner using FIRE. To train the model, we collected k-space data from ECG-segmented cine images from 343 patients undergoing clinical CMR. The training data were synthesized by discarding outer ky lines (i.e., reduced spatial resolution) and reconstructed using the same pipeline as the inline reconstruction. Evaluation One healthy subject and one patient with heart failure underwent a supine bike exercise cardiac MRI protocol. Highly accelerated real time cine short axis images were collected post-exercise with the following imaging parameters: SSFP, two-fold PE undersampling, temporal resolution = 35 ms, spatial resolution = 1.9 × 1.9 mm2. Two heart beats were needed for a single slice. The first was used to establish steady state and estimate the RR time interval, while the second for image acquisition lasting the calculated RR interval . Blurry images (i.e., due to 2-fold PE undersampling) resulting from vendor-provided 7-fold CS reconstruction were deblur using REGAIN. Then, DENT was applied to enable 2-fold gain in temporal resolution (Fig. 3).Results
Real time cine imaging post-exercise during a single heart beat per slice was achieved using proposed techniques. This was demonstrated at heart rates of 120 bpm in a healthy subject (Fig. 4) and 85 bpm in a patient (Fig. 5).Conclusion
We developed a highly accelerated high-frame-rate real-time cine for exercise cardiac MRI that was reconstructed sequentially with CS, REGAIN and DENT. Our approach enabled single-beat imaging with spatiotemporal resolution 1.9 × 1.9 mm2, 16 ms.Acknowledgements
No acknowledgement found.References
No reference found.Figures
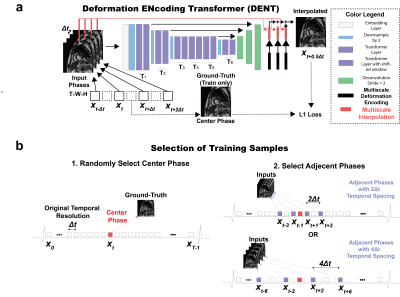
Figure 1. Deformation ENcoding Transformer (DENT). a) DENT works in image domain and generates
forward/backwards mappings describing underlying deformation in
multiple cardiac phases. Such deformation were used to synthesize images with
high framerate. DENT used input T = 4 sequential cine images of size W × H
collected with temporal resolution ∆t
at t - ∆t, t, t + ∆t, t + 2∆t. Output was interpolated image with equivalent time t + 0.5∆t. b) Center-phase (ground-truth) xt was
randomly selected. Then, 4 phases adjacent to xt were
selected as inputs with equally probably patterns 2∆t or 4∆t.
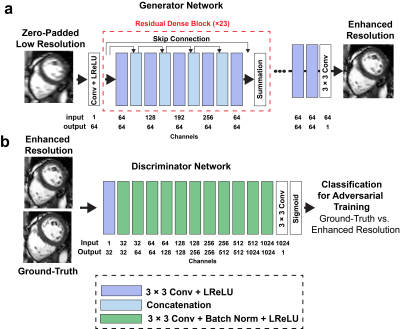
Figure 2. Resolution
Enhancement Generative Adversarial Inline Network (REGAIN). REGAIN acceleration prescribed 2-fold reduced phase-encode (PE). Image was reconstructed with zero-padding to create an image with full resolution but significant blurring. Then, REGAIN enhanced resolution, analogous to collecting a full-resolution image. The generator of REGAIN enhances the image resolution, and
the discriminator classifies the generator's output and high-resolution image
for the adversarial training. REGAIN was seamlessly integrated with the scanner using FIRE.
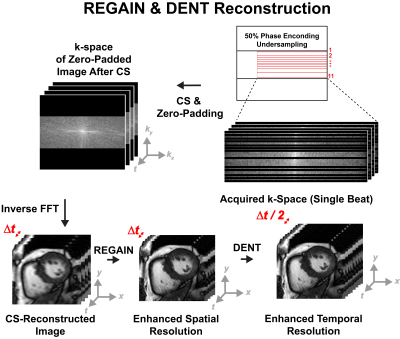
Figure 3. REGAIN and
DENT image reconstruction workflow. Data collection started after a dummy heart beat to take magnetization to steady-state b) k-space data were collected using an incoherent random 7-fold undersampling and reconstructed using CS with temporal resolution (TR) = 31.2 ms. REGAIN was used to deblur images (i.e., due to 2-fold reduced spatial resolution along phase-encoding direction). DENT was applied to synthesizing images between each two subsequent phases, resulting in cine images with spatial resolution of 1.9 × 1.9 mm2 and TR = 16 ms.
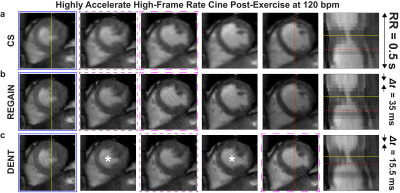
Figure 4. Exercise cardiac MRI in patient
with heart failure. a), Images were collected with compressed sensing (CS) rate = 7 and 2-fold
phase encoding (PE) undersampling. CS resulted in spatiotemporal resolution = 1.9 × 1.9 mm2,
35 ms. Images were blurry due low prescribed spatial resolution (i.e. 2-fold PE
undersampling). b), REGAIN processing
of CS-Recon images improves images sharpness. c) DENT was applied to REGAIN output to resulting in temporal
resolution of 15.5 ms. Boxes indicate corresponding cardiac phases across
methods. DENT-interpolated images indicated by asterisk.
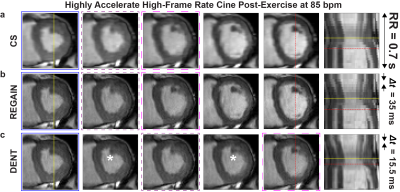
Figure 5. Exercise cardiac MRI in healthy
subject. Real time cine images were collected and reconstructed as
described in Figure 4. The healthy subject achieved a maximum heart rate
post-exercise of 159 bpm. The heart rate dropped to 120 bpm by the time the
cine slice was collected, which corresponded to an RR interval of 0.5 s.
Nevertheless, comparison of REGAIN and DENT images demonstrate left ventricular
relaxation occurs in short amount of time. DENT-interpolated images are indicated by asterisk.
DOI: https://doi.org/10.58530/2023/0339