0327
Anatomical-aware Siamese Deep Network for Prostate Cancer Detection on Multi-parametric MRI1Computer Science, University of California, Los Angeles, Los Angeles, CA, United States, 2Radiological Science, University of California, Los Angeles, Los Angeles, CA, United States, 3Radiology, The First Affiliated Hospital of China Medical University, Shenyang, China
Synopsis
Keywords: Prostate, Cancer, Machine Learning
The study aimed to build a deep-learning-based prostate cancer (PCa) detection model integrating the anatomical priors related to PCa’s zonal appearance difference and asymmetric patterns of PCa. A total of 220 patients with 246 whole-mount histopathology (WMHP) confirmed clinically significant prostate cancer (csPCa), and 432 patients with no indication of lesions on multi-parametric MRI (mpMRI) were included in the study. A proposed 3D Siamese nnUNet with self-designed Zonal Loss was implemented, and results were evaluated using 5-fold cross-validation. The proposed model that is aware of PCa-related anatomical information performed the best on both lesion-level detection and patient-level classification experiments.
Introduction
Multi-parametric MRI (mpMRI) is commonly used for prostate cancer (PCa) diagnosis1 . According to the Prostate Imaging Reporting and Data System, version 2 (PI-RADS)1, suspicious PCa in the transition zone (TZ) and peripheral zone (PZ) is diagnosed with different focus on different image components of mpMRI. Although there are deep-learning-based PCa detection models using mpMRI2-5, most of them ignore this PCa-related anatomical difference but treat all lesions identically2-4. A recent study considers zonal anatomical differences by stacking zonal masks to the input5. However, only modifying input might contribute limitedly towards optimal model convergence since no explicit constraints, like loss function, were additionally imposed. The PCa-related anatomical diagnostic priors could be further utilized and introduced as additional constraints to guide training and help conduct better model.Moreover, visual similarities existed between central zone (CZ), benign prostatic hyperplasia (BPH) and PCa, which may cause false-positive (FP) predictions6,7. BPH and CZ show symmetric patterns6,7, while PCa is commonly presented with asymmetric patterns8,9. These symmetric-related anatomical differences could be utilized to suppress FP predictions.
In this study, we proposed integrating PCa-related anatomical priors into our deep learning model to better detect clinically significant PCa (csPCa). Specifically, we proposed Siamese 3DnnUNet and a self-designed Zonal loss (ZL) that considers anatomical characteristics of PCa in different zones1,6-9.
Methods
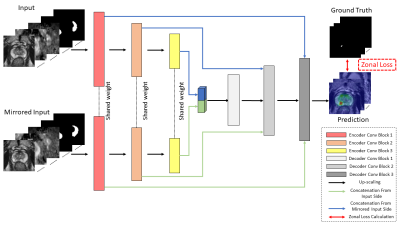
In the study, a total of 652 patients were included, consisting of 220 patients with 246 csPCa lesions, and 432 benign patients. mpMRI images were acquired from Siemens MRI machines, including T2WI, ADC, and high-B DWI images. The zonal masks were automatically generated by using another deep-learning-based prostate zonal segmentation model10 .Figure 1 shows the architecture of the proposed model, including Siamese 3DnnUNet and the ZL. The proposed network took input and mirrored input of 3D stacks of T2WI, ADC, high-B DWI, TZ, and PZ mask images and output the prediction probability map of csPCa.
As PCa should be treated differently in TZ and PZ1, zonal information was first provided by stacking the zonal masks as part of the network’s input (Figure 1). Moreover, we proposed to include a new hierarchical loss11, ZL, to further utilize PCa-related anatomical information as an additional constraint to guide model training. We assigned voxels of TZ and PZ lesions in lesion masks as labels [1, 1] and [0, 1]. This design was made to enable TZ lesions need extra T2WI for an improved diagnosis1. We defined the score map $$$P\in[0,1]^{H\times W\times 2}$$$, in which the two channels correspond to TZ and PZ. For each voxel $$$i$$$, the prediction probability vector $$$p_i=[p_v]_{v\in\{tz, pz\}}\in[0,1]^2$$$ in $$$P$$$ is given as: $$\begin{cases}p_{tz}&=min(s_{pz},s_{tz})\\1-p_{tz}&=1-s_{tz}\end{cases} \ \ \ \ \ \ \ \begin{cases}p_{pz}&=s_{pz}\\1-p_{pz}&=min(1-s_{tz}, 1-s_{pz})\end{cases}$$ where, $$$s_u$$$ is the prediction probability of voxels in zone $$$u$$$. An intuitive diagnostic interpretation of the ZL is that, highly suspicious TZ lesions and PZ lesions should share similar abnormalities on high-B DWI and ADC, but PZ lesions might not appear similarly to TZ lesions on T2WI1. The final representation of the ZL is: $$L(P)=\sum_{v\in\{tz,pz\}}-l_vlog(p_v)-(1-l_v)log(1-p_v)$$ where, $$$l_v$$$ for lesion voxels in zone $$$v(v\in\{TZ,PZ\}),l_v=0$$$ otherwise.
In addition, BPH and CZ can show similar imaging appearances to PCa1,6,7 . We implemented a Siamese Network12 (Figure 1) to guide the model to distinguish PCa from BPH and CZ as it can learn symmetric-related features effectively. We chose 3DnnUNet as the network backbone because it performs well on segmentation and detection tasks in medical imaging13 .
We compared our proposed model with other 3D deep learning models14-16, and also the baselines, 3DnnUNet without/with zonal masks, on csPCa detection and patient-level classification tasks. The model performance was tested via 5-fold cross-validation. The lesion-level csPCa detection and patient-level classification performances were evaluated by Free-response ROC (FROC) and ROC analysis, respectively.
Results
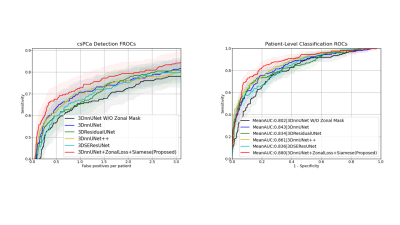
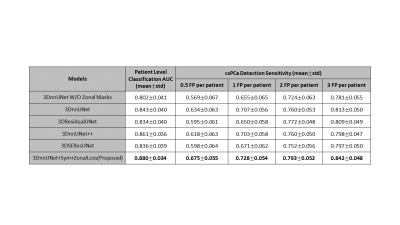
The comparisons are shown in Figure 2 and Figure 3. In the csPCa detection task, our proposed model achieved the highest sensitivities in every situation compared with 3DnnUNet++, 3DResidualUNet, and 3DSEResUNet14-16 . Compared with 3DnnUNet without zonal information, shown as 3DnnUNet W/O Zonal Mask, the proposed model showed improved sensitivities in all situations (Figure 3). Compared with 3DnnUNet with zonal masks but no additional PCa-related anatomical constraint, shown as 3DnnUNet, the proposed model also achieved superior sensitivities (Figure 3). In the patient-level classification task, our proposed model achieved the highest AUC of 0.880$$$\pm$$$0.034.Discussions
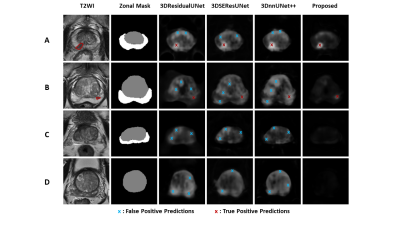
We proposed an anatomical-aware deep network composed of Siamese 3DnnUNet and a self-designed Zonal loss for csPCa detection. The results showed that integrating PCa-related anatomical priors into the construction of the deep learning model helped improve the model performance on both csPCa detection and patient-level classification tasks. We showed representative examples of lesion detection performances comparisons in Figure 414-16 . Compared with other models14-16 , the proposed model conducted less FP detections with same TP predictions on all cases. It also performs better on distinguishing patterns between PCa and symmetric abnormalities caused by BPH and CZ compared with other models14-16.Conclusions
In conclusion, we showed the proposed anatomical-aware prostate cancer detection deep network achieved accurate performance on csPCa detection and patient-level classification tasks. The integration of anatomical priors regarding symmetric patterns related to BPH and CZ, and diagnostic-related hierarchical Zonal Loss design helped improve the model performances on both lesion detection and patient-level classification tasksAcknowledgements
This HIPAA-compliant study was approved by the review board of our local institute with a waiver of informed consent. This work was supported by the National Institutes of Health (NIH) R01-CA248506 and funds from the Integrated Diagnostics Program, Department of Radiological Sciences & Pathology, David Geffen School of Medicine at UCLA.References
1. Weinreb JC, Barentsz JO, Choyke PL, et al. PI-RADS Prostate Imaging - Reporting and Data System: 2015, Version 2. European Urology. 2016;69(1):16-40
2. Cao R, Zhong X, Afshari S, et al. Performance of Deep Learning and Genitourinary Radiologists in Detection of Prostate Cancer Using 3-T Multiparametric Magnetic Resonance Imaging. J Magn Reson Imaging. 2021;54(2):474-483
3. Li D, Han X, Gao J, et al. Deep Learning in Prostate Cancer Diagnosis Using Multiparametric Magnetic Resonance Imaging With Whole-Mount Histopathology Referenced Delineations. Front Med (Lausanne). 2022;8:810995.
4. Iqbal S, Siddiqui GF, Rehman A, et al. Prostate Cancer Detection Using Deep Learning and Traditional Techniques. IEEE Access. 2021;9:27085-27100.
5. Hosseinzadeh M, Saha A, Brand P, et al. Deep learning-assisted prostate cancer detection on bi-parametric MRI: minimum training data size requirements and effect of prior knowledge. Eur Radiol. 2022;32(4):2224-2234.
6. Yu J, Fulcher AS, Turner MA, et al. Prostate cancer and its mimics at multiparametric prostate MRI. Br J Radiol. 2014; 87(1037): 20130659.
7. Panebianco V, Barchetti F, Barentsz J et al. Pitfalls in Interpreting mp-MRI of the Prostate: A Pictorial Review with Pathologic Correlation. Insights Imaging. 2015;6(6):611-30.
8. Smith RA, Mettlin CJ, Eyre H. Cancer screening and early detection. Holland-Frei cancer medicine. 6th edition. Hamilton, Canada: BC Decker; 2003.
9. Barentsz JO, Richenberg J, Clements R et al. European Society of Urogenital Radiology. ESUR prostate MR guidelines 2012. Eur Radiol. 2012;22(4):746-57.
10. Hung ALY, Zheng H, Miao Q, et al. CAT-Net: A Cross-Slice Attention Transformer Model for Prostate Zonal Segmentation in MRI. IEEE Trans Med Imaging. 2022.
11. Li L, Zhou T, Wang W et al. Deep Hierarchical Semantic Segmentation. in 2022 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), New Orleans, LA, USA, 2022.
12. Chicco D. Siamese Neural Networks: An Overview. In: Artificial Neural Networks. Methods in Molecular Biology. 2021;2190
13. Isensee F, Jaeger PF, Kohl SAA et al. nnU-Net: a self-configuring method for deep learning-based biomedical image segmentation. Nat Methods 18. 2021;203-211.
14. Zhou Z, Siddiquee MMR, Tajbakhsh N et al. UNet++: A Nested U-Net Architecture for Medical Image Segmentation. Deep Learn Med Image Anal Multimodal Learn Clin Decis Support. 2018;11045:3-11.
15. Zhang Z, Liu Q and Wang Y. Road Extraction by Deep Residual U-Net, in IEEE Geoscience and Remote Sensing Letters; 2018;15(5):749-753.
16. Hu J, Shen L and Sun G, Squeeze-and-Excitation Networks, 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), 2018;7132-7141
Figures