0322
Investigating tissue microstructure of prostate cancer using Linear Multi-scale Modeling of diffusion MRI data
Barbara Daria Wichtmann1, Niklas Westhoff2, Cleo-Aron Weis3, Ralph Strecker4, Thorsten Feiweier5, Steffen Albert6,7, Moritz Wolter8, Frank Zöllner6,7, Bettina Baeßler9, Aapo Nummenmaa10, Qiuyun Fan10,11, Susie Huang10,12, and Ulrike Attenberger1
1Diagnostic and Interventional Radiology, University Hospital Bonn, Bonn, Germany, 2Department of Urology and Urosurgery, Medical Faculty Mannheim, Heidelberg University, Mannheim, Germany, 3Institute of Pathology, University Hospital Heidelberg, Heidelberg, Germany, Heidelberg, Germany, 4EMEA Scientific Partnerships, Siemens Healthcare GmbH, Erlangen, Germany, 5MR Application Development, Siemens Healthcare GmbH, Erlangen, Germany, 6Computer Assisted Clinical Medicine, Medical Faculty Mannheim, Heidelberg University, Mannheim, Germany, 7Mannheim Institute for Intelligent Systems in Medicine, Medical Faculty Mannheim, Heidelberg University, Mannheim, Germany, 8High Performance Computing & Analytics Lab, University Bonn, Bonn, Germany, 9Institute of Diagnostic and Interventional Radiology, University Hospital Würzburg, Würzburg, Germany, 10A. A. Martinos Center for Biomedical Imaging, Department of Radiology, Massachusetts General Hospital, Charlestown, MA, United States, 11Department of Biomedical Engineering, College of Precision Instruments and Optoelectronics Engineering, Tianjin University, Tianjin, China, 12Harvard-MIT Division of Health Sciences and Technology, Massachusetts Institute of Technology, Cambridge, MA, United States
1Diagnostic and Interventional Radiology, University Hospital Bonn, Bonn, Germany, 2Department of Urology and Urosurgery, Medical Faculty Mannheim, Heidelberg University, Mannheim, Germany, 3Institute of Pathology, University Hospital Heidelberg, Heidelberg, Germany, Heidelberg, Germany, 4EMEA Scientific Partnerships, Siemens Healthcare GmbH, Erlangen, Germany, 5MR Application Development, Siemens Healthcare GmbH, Erlangen, Germany, 6Computer Assisted Clinical Medicine, Medical Faculty Mannheim, Heidelberg University, Mannheim, Germany, 7Mannheim Institute for Intelligent Systems in Medicine, Medical Faculty Mannheim, Heidelberg University, Mannheim, Germany, 8High Performance Computing & Analytics Lab, University Bonn, Bonn, Germany, 9Institute of Diagnostic and Interventional Radiology, University Hospital Würzburg, Würzburg, Germany, 10A. A. Martinos Center for Biomedical Imaging, Department of Radiology, Massachusetts General Hospital, Charlestown, MA, United States, 11Department of Biomedical Engineering, College of Precision Instruments and Optoelectronics Engineering, Tianjin University, Tianjin, China, 12Harvard-MIT Division of Health Sciences and Technology, Massachusetts Institute of Technology, Cambridge, MA, United States
Synopsis
Keywords: Prostate, Cancer
Linear Multi-scale Modeling (LMM) is an advanced diffusion-weighted imaging(DWI) technique that uses multi-shell, multi-diffusion-time DWI data to estimate tissue microstructure parameters, including volume fractions of restricted and hindered water compartments over a range of length scales and orientation distribution information. Here,we apply the LMM framework to characterize prostate cancer(PCa) lesions and correlate our results with histology. Within the histopathologically proven cancerous lesions we observed a significantly increased fraction of restricted diffusion, particularly within the 2μm and 7μm sized water compartments. LMM may enable the development of distinct diffusion microstructural signatures of PCa to facilitate diagnosis of clinically significant PCa lesions.Introduction
Diffusion-weighted imaging (DWI) is an integral part of multiparametric magnetic resonance imaging (mpMRI) for the diagnosis and prognosis of prostate cancer (PCa)1. Linear Multi-scale Modeling2 (LMM) is a recently developed advanced DWI technique that uses multi-shell, multi-diffusion-time DWI data to estimate tissue microstructure parameters, including volume fractions of restricted and hindered water compartments over a range of length scales and orientation distribution information. LMM has shown promise for identifying distinct diffusion microstructural signatures of pathology compared to healthy tissue3. In this work, we apply the LMM framework to characterize PCa-lesions and correlate our results with histology.Methods
Data acquisition: With ethics committee approval, 26 patients (mean age 67.7 +/- 4.9 years standard deviation) diagnosed with intermediate-risk adenocarcinoma of the prostate who were scheduled for radical prostatectomy were enrolled in this study. Examinations were performed on a clinical 3T scanner equipped with diffusion gradients up to 45mT/m (MAGNETOM Skyra; Siemens Healthcare, Erlangen, Germany) with the built-in spine matrix coil and the standard body matrix coil. Axial 2.7-mm isotropic resolution diffusion-weighted spin echo EPI images were acquired using a research application. The following parameters were used: δ/Δ=18/30 with b-values of 150s/mm2, 400s/mm2, 800s/mm2, and δ/Δ=18/60ms with b-values of 50s/mm2, 400s/mm2, 800s/mm2, 1500s/mm2, 2000s/mm2, TE/TR=91/4700ms, GRAPPA acceleration factor R=2. Diffusion gradients were applied in non-collinear directions (increasing with each q-shell up to 30 directions) with interspersed b=0 images every 10 directions. High-resolution axial, sagittal and coronal T2-weighted images of the prostate were also acquired. Total acquisition time was approximately 40min. Following prostatectomy, the resected specimen was processed with standard pathology and H&E staining.LMM analysis: Following preprocessing including DWI denoising using the MRtrix3 package 4, spherical harmonics expansion of order 6 with Laplace-Beltrami regularization5 (λ=0.006) was used to interpolate the diffusion signal on each q-shell. The linear multi-scale forward model of different sized restricted and hindered diffusion compartments was obtained by concatenating two spectra of response functions: 1) a non-Gaussian diffusion response function6 for water restricted inside cylindrical structures and 2) a Gaussian diffusion response function7 for hindered water and free water diffusion. For a more compact and efficient linear implementation we parameterized the orientation distribution of the hindered and restricted compartments with a set of order 4 and 6 spherical harmonics, respectively. To obtain the orientation distribution functions and corresponding volume fractions, the multi-scale deconvolution inverse problem was solved by standard least-squares estimation with Tikhonov regularization.
Segmentation: A convolutional neural network was used to automatically segment prostate zones on the axial high-resolution T2-weighted images8. Furthermore, lesions were segmented on the axial high-resolution T2-weighted images by an experienced radiologist with 5 years of experience. Registration of the DWI and T2-weighted images was performed using SimpleITK9,10.
Results
Voxel-wise estimated volume fractions of the different sized water compartments were clearly distinguishable between the peripheral zone and central/transitional zone (Figure 1). Within the histopathologically proven cancerous lesions we observed a significantly increased fraction of restricted diffusion, particularly within the 2μm and 7μm sized water compartments (Figure 1 and 2).Discussion
LMM enables detailed microstructural tissue characterization by separating orientation distributions of restricted and hindered diffusion water compartments over a range of length scales. Studies have shown that the median cell diameter of circulating tumor cells is 10.3μm which is slightly higher than our estimates11. While our results seem realistic, it is to note that higher diffusion gradients will most likely improve accuracy and increase our certainty in cell diameter estimation12.The estimation of restricted and hindered volume fractions and compartment sizes may provide insight into the distinct microstructural features of PCa lesions. A recently published study on time-dependent DWI for microstructural mapping of PCa found higher Gleason grades to be correlated with a higher degree of diffusion restriction and smaller cell diameters13. Considering the risk of overdiagnosis and overtherapy, LMM might particularly aid in the diagnosis of clinically significant PCa lesions. As this study only included patients with diagnosed intermediate-risk adenocarcinoma that were scheduled for radical prostatectomy further investigation in this regard is required.
Conclusion
In its implementation as a linear inverse problem, LMM offers a powerful, yet easy to compute analysis framework that enables a detailed characterization of tissue microstructure and orientation distribution. LMM may enable the development of distinct diffusion microstructural signatures of PCa to facilitate diagnosis of clinically significant PCa lesions.Acknowledgements
This research project is part of the Research Campus M2OLIE and funded by the German Federal Ministry of Education and Research (BMBF) within the Framework “Forschungscampus: public-private partnership for Innovations” under the funding code 13GW0388A.References
- Wichtmann BD et al.: Multiparametric MRI in the Diagnosis of Prostate Cancer: Physical Foundations, Limitations, and Prospective Advances of Diffusion-Weighted MRI. Rofo. 2021 Apr;193(4):399-409. English. doi: 10.1055/a-1276-1773. Epub 2020 Dec 10. PMID: 33302312.
- Wichtmann BD et al.: Linear multi-scale modeling of diffusion MRI data: A framework for characterization of oriented structures across length scales. Hum Brain Mapp. In press.
- Wichtmann BD et al.: Investigating microstructural signatures for low-grade gliomas using Linear Multi-scale Modeling of diffusion MRI data. Proceedings ISMRM 2017: 3362.
- Tournier JD, et al.: MRtrix3: A fast, flexible and open software framework for medical image processing and visualisation. NeuroImage, 202 (2019), pp. 116–37.
- Descoteaux M et al.: Regularized, fast, and robust analytical Q-ball imaging. Magn Reson Med. 2007 Sep;58(3):497-510. doi: 10.1002/mrm.21277. PMID: 17763358.
- van Gelderen P et al.: Evaluation of restricted diffusion in cylinders. Phosphocreatine in rabbit leg muscle. J Magn Reson B. 1994 Mar;103(3):255-60. doi: 10.1006/jmrb.1994.1038. PMID: 8019777.
- White NS et al.: Probing tissue microstructure with restriction spectrum imaging: Histological and theoretical validation. Hum Brain Mapp. 2013 Feb;34(2):327-46. doi: 10.1002/hbm.21454. Epub 2012 Jan 16. PMID: 23169482; PMCID: PMC3538903.
- Meyer A et al.: Towards patient-individual PI-Rads v2 sector map: CNN for automatic segmentation of prostatic zones from T2-weighted MRI. IEEE 16th International Symposium on Biomedical Imaging (ISBI 2019): 696-700.
- Lowekamp BC et al.: The Design of SimpleITK. Front Neuroinform. 2013 Dec 30;7:45. doi: 10.3389/fninf.2013.00045. PMID: 24416015; PMCID: PMC3874546.
- Yaniv Z et al.: SimpleITK Image-Analysis Notebooks: a Collaborative Environment for Education and Reproducible Research. J Digit Imaging. 2018 Jun;31(3):290-303. doi: 10.1007/s10278-017-0037-8. Erratum in: J Digit Imaging. 2019 Sep 4;: PMID: 29181613; PMCID: PMC5959828.
- Mendelaar PAJ et al.: Defining the dimensions of circulating tumor cells in a large series of breast, prostate, colon, and bladder cancer patients. Mol Oncol. 2021 Jan;15(1):116-125. doi: 10.1002/1878-0261.12802. Epub 2020 Oct 4. PMID: 32949099; PMCID: PMC7782084.
- Huang SY et al.: The impact of gradient strength on in vivo diffusion MRI estimates of axon diameter. Neuroimage. 2015 Feb 1;106:464-72. doi: 10.1016/j.neuroimage.2014.12.008. Epub 2014 Dec 9. PMID: 25498429; PMCID: PMC4285777.
- Wu D et al.: Time-Dependent Diffusion MRI for Quantitative Microstructural Mapping of Prostate Cancer. Radiology. 2022 Jun;303(3):578-587. doi: 10.1148/radiol.211180. Epub 2022 Mar 8. PMID: 35258368.
Figures
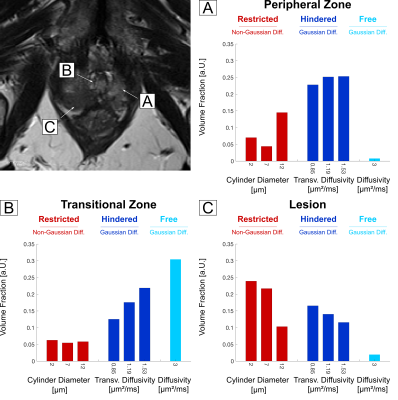
Voxel-wise estimated volume fractions of the different sized water compartments were clearly distinguishable between the peripheral zone and central/transitional zone with a particularly higher degree of free diffusion within the transitional zone. Within the histopathologically proven cancerous lesions we observed a significantly increased fraction of restricted diffusion, particularly within the 2μm and 7μm sized water compartments.
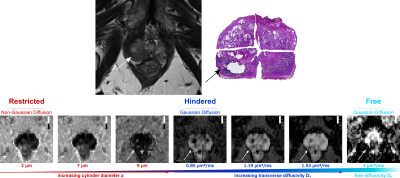
Axial T2-weighted image of a prostate with a PI-RADS 5 lesion in the right posterolateral peripheral zone and suspected perineural invasion (upper row left). Histophatology revealed adenocarcinoma with a Gleason-score of 4+5=9 and confirmed infiltration of the perineural sheath (upper row right). The lower row shows the corresponding volume fraction maps with increasing diffusion length scale from left to right. These maps visualize the contribution to the total signal per diffusion length scale (bright voxels - large contribution, dark voxels - little to no contribution).
DOI: https://doi.org/10.58530/2023/0322