0286
Metabolic Imaging of Alzheimer’s Disease Related Inflammation Using Deuterium MRI
Matthew E Merritt1, Mario Chang1, Tara Hawkinson1, Anna Rushin1, Vikram Kodibagkar2, James Bankson3, and Ramon Sun1
1Biochemistry and Mol. Bio., University of Florida, Gainesville, FL, United States, 2School of Biological and Health Systems Engineering, Arizona State University, Tempe, AZ, United States, 3Imaging Physics, MD Anderson Cancer Center, Houston, TX, United States
1Biochemistry and Mol. Bio., University of Florida, Gainesville, FL, United States, 2School of Biological and Health Systems Engineering, Arizona State University, Tempe, AZ, United States, 3Imaging Physics, MD Anderson Cancer Center, Houston, TX, United States
Synopsis
Alzheimer’s Disease (AD) is characterized by increased inflammation and increased glucose utilization as detected by FDG-PET. Using a new deuterium MRI method based on a [D7]glucose tracer, we can image the brain of a mouse model of AD with in plane resolution approaching 1 mm. Mass spectrometry imaging of the same brains shows characteristic changes in protein associated glycans that correlate well with immunohistochemical staining for inflammation. We believe this new research pipeline can provide powerful new insights into AD pathophysiology.
Introduction
Alzheimer’s disease (AD) is a tremendous burden to the healthcare system, with an estimated 6 million US citizens afflicted by the disease in 2020. AD is characterized by progressive loss of cognitive function, and is phenotypically recognized by increased deposition of protein fibrils in the brain. Furthermore, the AD brain is characterized by significant neuroinflammation related to endoplasmic reticulum (ER) stress, improper protein glycosylation and subsequent protein misfolding. Glucose uptake as measured by 18F-deoxyglucose positron emission tomography (FDG-PET) is commonly used to assess AD in vivo, as the AD brain is known to consume less glucose. Recently we have shown that deuterium magnetic resonance imaging (DMRI) using a [D7]glucose contrast agent is sensitive to glycolysis through the generation of partially deuterated water (HDO) and lactate, providing a readout not only of glucose uptake but also its downstream metabolism. With the advent of matrix-assisted laser desorption ionization-mass spectrometry imaging (MALDI-MSI), it is now possible to assess glucose incorporation into the brain as glycogen or due to protein post-translational modifications, providing an orthogonal readout of glucose metabolism that is orthogonal to glucose oxidation for energy production. Hypothesis: A combined DMRI-MALDI imaging approach will determine if the decreased glucose uptake in AD is related to increased neuroinflammation and subsequent damage energy metabolism.Methods
MRI. A common murine model of AD is the 5xFAD mouse. 5xFAD mice and age matched controls were imaged following tail vein injection of [D7]glucose at a dosage of 1.95 g/kg. Imaging was carried out at 11 T using a Bruker Avance III HD imaging console. A 2.6 cm 2H saddle coil was placed over the mouse brain, and the whole animal was localized within a 4 cm 1H saddle coil used for prospective imaging. Background 2H images were acquired using a fast low-angle single shot (FLASH) sequence. Upon injection, unlocalized 2H spectroscopy was used to monitor the arrival of the substrate in the brain. At approximately 8 minutes post-injection, a two-point Dixon (2PD) multi-echo gradient sequence was used to image HDO versus glucose peaks. Alternatively, a 2D-chemical shift imaging sequence was used.MALDI-MSI
Brains of mice were removed and drop fixed in 10% neutral buffered formalin (NBF) for 24h. The fixative was switched to 70% EtOH after 24h, and after another 24h the tissue was embedded in paraffin blocks. FFPE slides were cut at 4uM for both immunohistochemical (IHC) and MALDI analysis. MALDI slides were dewaxed prior to antigen retrieval. Slides were then sprayed with PNGaseF to cleave glycans. After a 4h incubation, the slides were then sprayed with alpha-cyano hydroxycinnamic acid (CHCA) and immediately ran on a Water’s Synapt G2 Si MALDI-MSI. Data was exported to HDI (Water’s corporation) where frontal cortex regions of interest were circled, and abundance values exported to a CSV file for the generation of graphs.
Results
The DMRI method allows for high in-plane spatial resolution in a reasonable time frame (~13 minutes to acquire 32*32 resolution over a 35 mm FOV) (Figure 1). The 2PD method allows images of HDO and glucose to be reconstructed, allowing estimates of uptake and energy metabolism to be produced concurrently (Figure 2). MALDI imaging of the mouse brain indicated profound dysregulation of protein glycosylation in the 5xFAD mouse (Figure 3). Glial activation as a marker for neuroinflammation was determined using immunohistochemical markers such as glial fibrillary acidic protein (GFAP) and ionized calcium binding adaptor molecule 1 (IBA1). IHC analyses was performed on WT, and 5xFAD mouse models to elucidate the extent of microglial activation (Figure 4).Conclusions
Neuroinflammation is particularly difficult to visualize with any in vivo imaging method. Our proposed research pipeline provides not only in vivo estimates of glucose uptake and utilization, but also enables a spatial readout of metabolic changes via MALDI imaging. Future experiments will directly correlate glucose oxidation with local inflammation in the 5xFAD mouse with an eye towards clinical translation of the MRI method.Acknowledgements
the authors acknowledge funding from NIH R01 DK132254 and EB032376.References
No reference found.Figures
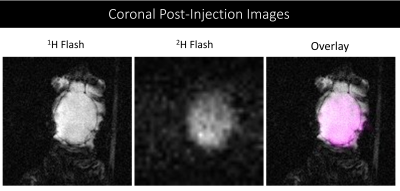
Figure 1. A. FLASH 1H coronal image
for anatomical imaging of the normal mouse. B. FLASH 2H image acquired from 8-15
minutes after the [D7]glucose injection. C. Overlay of images shows a uniform
utilization of glucose in the normal brain.
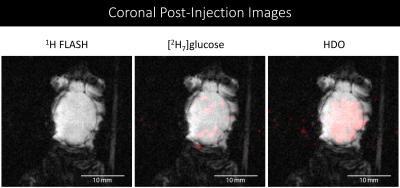
Figure 2. A. Coronal 2PD images of the glucose
and components acquired from 15-28 minutes post injection. The majority of the
signal is derived from metabolically derived HDO.
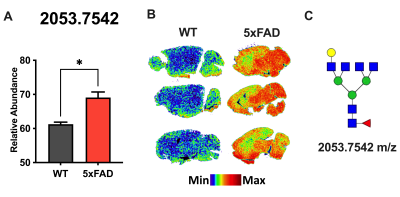
Figure 3. Mouse AD brains are
hyperglycosylated. (A) The relative abundance of glycan Hex4dHex1HexNAc6
(m/z: 2053) in WT and 5xFAD mouse models. Data shown represents the mean
+/- the SEM of n=3 per group. *P < 0.05; unpaired, two-tailed T test. (B) Spatial distribution of Hex4dHex1HexNAc6 in three
biological replicates of WT (left), and 5xFAD (right) mouse
brains with min/max color gradient represented below images. (C) Graphical representation
of glycan structure: blue square, N-acetylglucosamine, red triangle, fucose, green circle, mannose, and yellow circle, galactose.
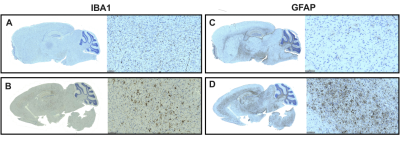
Figure 4. AD mouse brains reveal
increases in markers for glial activation and neuroinflammation A-B. Left:
Whole brain image for ionized calcium binding adapter 1 (IBA1) in WT (A), and 5xFAD (B)
mouse models. Right: zoomed-in frontal cortex. C-D. Left: Whole brain image for glial
fibrillary acidic protein (GFAP) in WT (C) and 5xFAD (D) mouse
models. Right: zoomed-in frontal cortex.
DOI: https://doi.org/10.58530/2023/0286