0250
The Brainstem Navigator: a toolkit to investigate brainstem nuclei structure, function, and connectivity in living humans1Brainstem Imaging Laboratory, Department of Radiology, Athinoula A. Martinos Center for Biomedical Imaging, Massachusetts General Hospital and Harvard Medical School, Boston, MA, United States, 2Division of Sleep Medicine, Harvard University, Boston, MA, United States, 3Research Center “E. Piaggio”, School of Engineering, University of Pisa, Pisa, Italy, 4Medical Physics Section, Department of Biomedicine and Prevention, Faculty of Medicine, Tor Vergata University of Rome, Rome, Italy, 5Escuela Nacional de Estudios Superiores Unidad Juriquilla, Universidad Nacional Autónoma de México, Queretaro, Mexico
Synopsis
Keywords: Software Tools, Brain, Brainstem atlas, segmentation, functional and structural connectome, 7 Tesla MRI, 3 Tesla MRI
Brainstem nuclei are deep gray matter regions involved in vital functions such as arousal/sleep/motor/sensory/autonomic/nociceptive/limbic/sensory function. Due to their small size and limited MRI contrast, these nuclei are difficult to visualize in conventional imaging of living humans. We generated and released the Brainstem Navigator toolkit, which enables the automatic identification of brainstem nuclei location in conventional and advanced MRI of living humans. It includes in-vivo probabilistic atlas labels for 31 brainstem nuclei generated by multi-contrast 7 Tesla MRI. We also developed coregistration routines optimized for the brainstem in 3 Tesla and 7 Tesla MRI data in health and disease.
Introduction
Brainstem nuclei are deep gray matter regions involved in vital functions such as arousal/sleep, motor, sensory, autonomic, nociceptive, limbic, and sensory function1. Due to their small size and limited MRI contrast, these nuclei are difficult to visualize in conventional imaging of living humans, and extrapolations from 2D post-mortem brainstem atlases are often used to identify their location in in-vivo MRI, with limited precision.Purpose
To generate and publicly release the Brainstem Navigator, a toolkit that enables the automatic identification of brainstem nuclei location in conventional (e.g. 3 Tesla) and advanced (e.g. 7 Tesla) MRI of living humans. The first goal was to create a probabilistic and deformable 3D atlas of 31 brainstem nuclei by 7 Tesla multi-contrast MRI. The second goal was to develop a tutorial suite of coregistration routines for precise alignment of the brainstem nuclei atlas labels to diffusion and functional MRI.Methods
Atlas generation and validation: For details see2-7. In brief, multi-contrast 1.1mm-isotropic-resolution echo-planar images were acquired at 7 Tesla in 12 subjects (mean±s.e age: 28±1 years, 6m/6f). After preprocessing and precise coregistration of T2-weighted and diffusion fractional anisotropy (FA) images to stereotactic space, semi-automatic and/or manual segmentations of brainstem nuclei were performed. Spatial overlap across subjects was then computed to yield probabilistic atlas labels of 31 brainstem nuclei, for a total of 76 labels including left/right/medial nuclei and multiple sub-nuclei. The nuclei atlas labels were validated by internal consistency or/and intra-rater agreement, volume comparison to post-mortem literature and histology. The atlas was developed in both 1mm-isotropic-resolution Illinois-Institute-of-Technology (IIT, 72 subjects) stereotactic space to facilitate its use in diffusion MRI studies, and in Montreal-Neurological-Institute (MNI ICBM,152 subjects) stereotactic space for functional MRI studies. Tutorial: Coregistration routines: To align user’s diffusion data to the brainstem nuclei atlas, we recommend high dimensional diffeomorphic bivariate non-linear registrations (e.g. ANTs command antsRegistration, registrations parameters detailed in8-9) of native FA/T2-weighted images to IIT FA/T2-weighted templates, either subject by subject, or via an optimal group template (e.g. ANTs script antsMultivariateTemplateConstruction2.sh). Note that the non-diffusion weighted “b0” image is a T2-weighted image. To align functional MRI data to the brainstem nuclei atlas in MNI space (for details see10-11), first, a subject-by-subject boundary based affine or non-linear registration to the anatomical T1-weighted MRI (e.g. MPRAGE) is computed, and applied to the native functional MRI; second, a univariate non-linear transformation between the native T1-weighted MRI and the T1-weighted MNI template is computed, and applied to the functional MRI brought to native T1-weighted MRI space. The second step can be performed (e.g. using ANTs) either subject-by-subject, or via an optimal group template (the latter can also be used to streamline mask computation). Brain masking for coregistration routines: A crucial step for precise brainstem coregistration is masking of areas outside the brain, which might be present in native images and absent in stereotactic templates. This includes cropping the inferior slices covering the spinal cord, removing scalp and non brain signals (e.g. running FSL bet on bias field corrected images), and manual editing of the brain mask, especially to remove areas anterior to the pons and medulla, which include for instance basilar vessels. Example application of the Brainstem Navigator to 7 Tesla and 3 Tesla MRI: The Brainstem Navigator atlas and coregistration routines were applied to 7 Tesla and 3 Tesla functional and diffusion data acquired on 20 healthy subjects (age 29.5 ± 1.1 years, 10m/10f). Resting state functional MRI connectivity and diffusion-based tractography were computed for 31 brainstem nuclei towards a parcellation of the whole brain, and resulting connectomes were compared across 7 Tesla and 3 Tesla scanners (see8-11 for details).Results
The brainstem Navigator toolkit has been released at https://www.nitrc.org/projects/brainstemnavig/ on January 2022, and it has been downloaded more than 645 times. In Figure 1, we show a 3D rendering of the in-vivo probabilistic atlas of 31 brainstem nuclei mainly involved in arousal/sleep/motor/autonomic/nociceptive/limbic/sensory function. In Figures 2-3, we show schematics of recommended pipelines for diffusion and functional MRI, respectively, with examples that demonstrate coregistration quality. In Figure 4, we show example uses of the Brainstem Navigator to map diffusion and functional based connectomes in living humans at 7 Tesla, and in Figure 5 translatability of the results to 3 Tesla MRI.Discussion and Conclusions
Our work demonstrates the feasibility of mapping the Brainstem Navigator atlas to 3 Tesla and 7 Tesla MRI data in health8-11 and disease7,12 by the use of precise coregistration routines optimized for the brainstem. Brain masking is crucial for precise brainstem coregistration, especially for coregistration of diffusion MRI and of T1-weighted MRI to stereotactic space. The estimated research/clinical user community size of the Brainstem Navigator toolkit is medium-large (1,000-5,000): this is based on a PubMed query resulting in 5,654 3 Tesla and 7 Tesla MRI studies of arousal/sleep, motor, sensory, autonomic/limbic/nociceptive function, connectivity and intervention (DBS, ablation, neurosurgery) that could be extended to the brainstem. We foresee that the Brainstem Navigator toolkit will facilitate investigation of brainstem structure and function in health and disease.Acknowledgements
Michael J. Fox Foundation; NIH (NIA-R01AG063982;NIDCD-R21DC015888;NIBIB-K01EB019474;NIBIB-P41EB015896); Massachusetts-General-Hospital Claflin-Distinguished-Scholar-Award; Harvard-University Mind-Brain-Behavior-Faculty-Award; Dr. Thorsten Feiweier for providing the diffusion sequence used in this study.
*M.G. García-Gomar and K. Singh are equally contributing authors.
References
1. Olszewski J and Baxter D. Cytoarchitecture of the human brain stem. Basel: Karger; 1954.
2. Bianciardi M, Toschi N, Edlow B L, et al. Toward an in vivo neuroimaging template of human brainstem nuclei of the ascending arousal, autonomic, and motor systems. Brain connectivity. 2015;5(10):597-607.
3. Bianciardi M, Strong C, Toschi N, et al. A probabilistic template of human mesopontine tegmental nuclei from in vivo 7 T MRI. Neuroimage. 2018;170:222-230.
4. García-Gomar MG, Strong C, Toschi N, et al. In vivo probabilistic structural atlas of the inferior and superior colliculi, medial and lateral geniculate nuclei and superior olivary complex in humans based on 7 Tesla MRI. Front Neurosci. 2019;13:764.
5. Singh K, Indovina I, Augustinack J C, et al. Probabilistic template of the lateral parabrachial nucleus, medial parabrachial nucleus, vestibular nuclei complex, and medullary viscero-sensory-motor nuclei complex in living humans from 7 Tesla MRI. Front Neurosci. 2020;13:1425.
6. Singh K, García-Gomar MG, Bianciardi M. Probabilistic atlas of the mesencephalic reticular formation, isthmic reticular formation, microcellular tegmental nucleus, ventral tegmental area nucleus complex, and caudal–rostral linear raphe nucleus complex in living humans from 7 Tesla magnetic resonance imaging. Brain Connect. 2021;11(8):613-623.
7. García‐Gomar MG, Videnovic A, Singh K, et al. Disruption of brainstem structural connectivity in REM sleep behavior disorder using 7 Tesla magnetic resonance imaging. Mov Disord. 2022;37(4):847-853.
8. Singh K, García-Gomar MG, Cauzzo S, et al. Structural connectivity of autonomic, pain, limbic, and sensory brainstem nuclei in living humans based on 7 Tesla and 3 Tesla MRI. Hum Brain Mapp. 2022;43(10):3086-3112.
9. García-Gomar MG, Singh K, Cauzzo S, Bianciardi M. In vivo structural connectome of arousal and motor brainstem nuclei by 7 Tesla and 3 Tesla MRI. Hum Brain Mapp. 2022;43(14):4397-4421.
10. Singh K, Cauzzo S, García-Gomar MG, et al. Functional connectome of arousal and motor brainstem nuclei in living humans by 7 Tesla resting-state fMRI. Neuroimage. 2022;249:118865.
11. Cauzzo S, Singh K, Stauder M, et al. Functional connectome of brainstem nuclei involved in autonomic, limbic, pain and sensory processing in living humans from 7 Tesla resting state fMRI. Neuroimage. 2022;250:118925.
12. Bianciardi M, Izzy S, Rosen BR, et al. Location of Subcortical Microbleeds and Recovery of Consciousness After Severe Traumatic Brain Injury. Neurology. 2021;97(2):e113-e123.
Figures
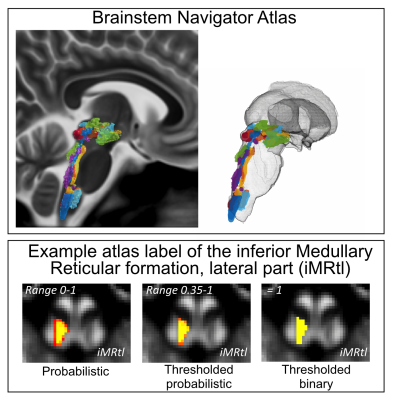
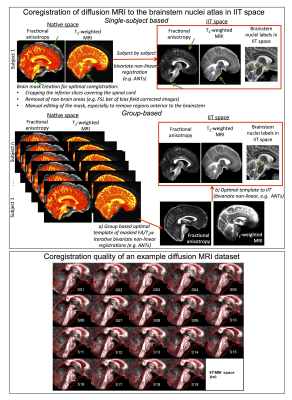
Figure 2 Top) Coregistration of diffusion MRI to the brainstem atlas in IIT space (subject-by-subject or group based). Masking out the scalp, cervical-spine and basilar vessels (green arrows) is crucial to achieve a good coregistration. To avoid time-consuming single-subject mask editing, a mask can be created on a group-based template of unmasked MRIs, and coregistered back to single-subject MRIs to generate single-subject masks. Bottom) Coregistration quality of an example diffusion MRI data set: the IIT template (red edges) is overlaid on single subject diffusion MRI8-9.
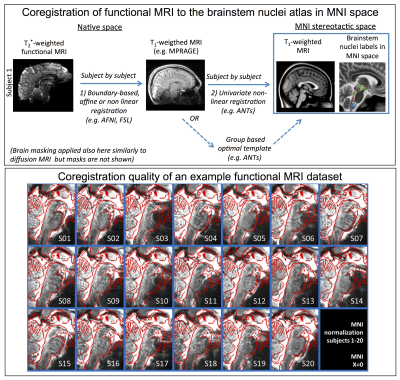
Figure 3 Top) Two-step coregistration procedure of functional MRI to the brainstem nuclei atlas in MNI space. 1) Boundary based affine or non-linear coregistration of native functional to T1-weighted MRI; notably, affine usually worked well for the cortex, and non-linear with mild deformations provided improvements for the brainstem; 2) Non-linear coregistration of native T1-weighted MRI to T1-weighted MNI template. Bottom) Coregistration quality of an example functional MRI data set: the MNI template is shown with red edges overlaid on single subject functional MRI10-11.
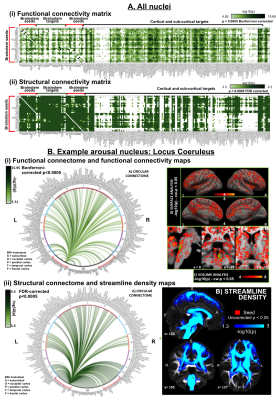
Figure 4 Application of the Brainstem Navigator atlas to map brainstem connectivity in living healthy humans using 1.1mm fMRI and 1.7mm diffusion 7 Tesla MRI8-11. A. Connectivity matrix of 18 brainstem nuclei seeds with cortical, sub-cortical and brainstem targets; functional connectivity resembled structural connectivity (r = 0.15, p = 5.5 10-38), yet it was denser with the cortex and sparser within the brainstem. B. (Left) Functional and structural connectomes of locus coeruleus, and (Right) its voxel-based functional connectivity map (top) and tract density map (bottom).
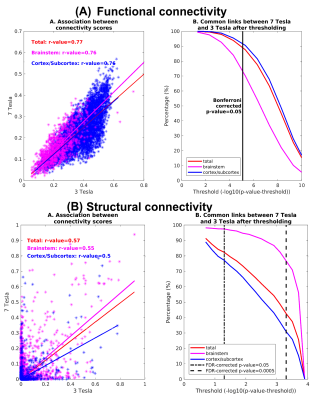
Figure 5 Translatability of 7 Tesla connectivity results to 3 Tesla 2.5mm-isotropic MRI8-11. Association and repeatability of links of functional (A) and (B) structural connectomes of 18 arousal and motor brainstem nuclei obtained at 3 Tesla compared to 7 Tesla MRI. The association between 3 Tesla and 7 Tesla connectomes was good, and was higher for the functional than the structural connectivity indices. The repeatability of links across scanners decreased with increasing the statistical threshold, and was above 75 % for p < 0.05 corrected for multiple comparisons.