0242
Automatic Rectal Tumor Segmentation and Extramural Venous Invasion Diagnosis based on Deep Learning and Radiomics Model1Institute of Science and Technology for Brain-Inspired Intelligence, Fudan University, Shanghai, China, 2Human Phenome Institute, Fudan University, Shanghai, China, 3Xiangya Hospital Central South University, Changsha, Hunan, China, 4Siemens Healthineers, Shanghai, China
Synopsis
Keywords: Machine Learning/Artificial Intelligence, Segmentation
This study developed an automatic diagnosis model for rectal cancer, which consists following steps: high-precision rectal tumor segmentation by Spatial Hybrid Network (SH-Net) and Adaboost Decision Tree based radiomics model to improve the diagnostic performance of extramural venous invasion (EMVI). The comparable diagnostic performance of the proposed model compared to the visual assessment by radiologists suggests the potential to help doctors with clinical diagnosis of EMVI.
Introduction
Extramural venous invasion (EMVI) is an independent predictor of local recurrence and distant metastases in rectal cancer (RC)1,2,3, which is traditionally diagnosed by the postoperative pathological specimen, namely preoperative EMVI (pEMVI). European Society for Medical Oncology recommend the assessment of MRI-based identification of EMVI (mrEMVI)4. Nevertheless, the assessment of mrEMVI is still challenging due to heterogeneous image quality, radiologists’ experience, and detection in very small vessels (≤ 3 mm)5. Therefore, there is an urgent need for an automatic, objective, sensitive method for EMVI evaluation.Methods
A retrospective study included 438 patients with rectal cancer who had T2WI images from a Siemens 1.5T scanner as well as pEMVI diagnostic report. These patients were randomly divided into a training cohort (332 patients: 130 pEMVI and 202 non-pEMVI) and a test cohort (106 patients: 52 pEMVI and 54 non-pEMVI). The Regions of interest (ROIs) were manually drawn on T2WI images using ITK-SNAP software (www.itksnap.org), including the circumferential margin, the rectal wall and the whole tumor. The proposed Spatial Hybrid Network (SH-Net) and Adaboost Decision Tree based radiomics (Ada-SH (max)) model consists of the following main steps (Figure 1): Automatic segmentation of rectal tumor from T2WI images based on SH-Net (Figure 2) with dilation post-processing which uses 10x10 structure element (kernel=10). We take advantage of 2D (high segmentation accuracy) and 3D (high smoothness of 3D organ contour) representations to design a spatial hybrid network to achieve more accurate segmentation through the fusion of multi-level feature maps (MLF). After auto-segmentation of tumor, radiomics features were extracted from the segmented tumor mask using Pyradiomics (https://github.com/AIM-Harvard/pyradiomics) and were dimensionally reduced using the Student’s t-test and Least Absolute Shrinkage and Selection Operator (LASSO) algorithm. Moreover, Synthetic Minority Oversampling Technique (SMOTE) was applied to adjust the imbalance of the data, and finally, AdaBoost Decision Tree algorithm was used to construct a classifier model for EMVI prediction. In addition, we compared our proposed model with radiologists’ assessment, radiomics model which uses manual ROIs (Ada-radiologist model), Ada-SH model which used the minimum delineation of predicted tumor segmentation without dilation (Ada-SH (min) model) to demonstrate the effectiveness of the proposed model. In terms of performance evaluation, the segmentation performance was evaluated by the DICE coefficient, and the performance of the radiomics analysis was assessed with respect to its receiver operating characteristics (ROC) curve, the area under ROC curve (AUC), accuracy, sensitivity, specificity, and f1-score.Results
The proposed Ada-SH model achieved a dice coefficient of 91.8% for the circumferential margin, 92.8% for the wall, and 79.5% for the tumor in the test cohort. The specific results are shown in Table 1. As shown in Figure 3 (a-e), Compared with some other comparative segmentation methods, the segmentation results of the proposed model are closest to the ground truth. In radiomics model building, 1145 features were extracted from tumor region segmented by the deep learning model and finally 53 features were retained to construct the radiomics signature after dimension reduction. The proposed Ada-SH (max) model achieved an AUC of 0.935, ACC of 0.840, sensitivity of 0.823 and specificity of 0.851 in the training cohort, and AUC of 0.854, ACC of 0.783, sensitivity of 0.807, and specificity of 0.859 in the test cohort. More importantly, we found that the Ada-SH model outperformed the radiologist's assessment in both the training and test cohorts in terms of AUC and sensitivity (Figure 3-f), demonstrating the feasibility of our model in clinical applications (Table 2). In the comparison of different ROIs, In the testing cohort, our Ada-SH (max) model suggests a better performance compared to Ada-SH (min) model (0.844), achieving no significant difference from Ada-radiologist model (0.863).Discussion
The results demonstrated the reliable performance of automatic segmentation of rectal tumor and diagnostic EMVI using Ada-SH (max) model based on T2WI images, and the proposed model yields increased AUC and sensitivity in comparison to radiologists’ diagnosis. Due to the thick-slice scanning causing severe inter-slice discontinuities of 3D MRI images, our SH-Net model can better balance inter-slice sparse information and intra-slice dense information to achieve higher segmentation accuracy. It can be proved that the tumor region segmented by our proposed SH-Net model achieved competitive results with the ROI manually labeled by radiologists in our radiomics model for EMVI diagnosis. In addition, since the EMVI of rectal cancer is determined by tumor cells in the blood vessels around the tumor, expansion operation is essential for the segmented regions to cover more valid information, which yields increased almost all the evaluation index in comparison to Ada-SH (min) model. Through training, the Ada-SH (max) model was capable of capturing those unique characteristics from T2WI images and distinguishing the EMVI of rectal tumor, resulting in superior performance in the segmentation and diagnosis for EMVI of rectal tumor, which is more accurate than radiologist diagnosis.Conclusion
This study implemented deep learning and radiomics analysis to auto-diagnose EMVI for rectal cancer patients. The superior performance both in the training cohort and test cohort indicated the proposed model may be reliable and valuable with high accuracy without surgery, which can greatly help doctors in clinical diagnosis and treatment.Acknowledgements
No acknowledgement found.References
- Zech C J. MRI of extramural venous invasion in rectal cancer: a new marker for patient prognosis?[J]. Radiology, 2018, 289(3): 686-687.
- Tudyka V, Blomqvist L, Beets-Tan R G H, et al. EURECCA consensus conference highlights about colon & rectal cancer multidisciplinary management: the radiology experts review[J]. European Journal of Surgical Oncology (EJSO), 2014, 40(4): 469-475.
- Betge J, Pollheimer M J, Lindtner R A, et al. Intramural and extramural vascular invasion in colorectal cancer: prognostic significance and quality of pathology reporting[J]. Cancer, 2012, 118(3): 628-638.
- Glynne-Jones R, Wyrwicz L, Tiret E, et al. Rectal cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up[J]. Annals of Oncology, 2017, 28: iv22-iv40.
- Bae J S, Kim S H, Hur B Y, et al. Prognostic value of MRI in assessing extramural venous invasion in rectal cancer: multi-readers’ diagnostic performance[J]. European radiology, 2019, 29(8): 4379-4388.
- Çiçek Ö, Abdulkadir A, Lienkamp S S, et al. 3D U-Net: learning dense volumetric segmentation from sparse annotation[C]//International conference on medical image computing and computer-assisted intervention. Springer, Cham, 2016: 424-432.
- Isensee F, Jaeger P F, Kohl S A A, et al. nnU-Net: a self-configuring method for deep learning-based biomedical image segmentation[J]. Nature methods, 2021, 18(2): 203-211.
Figures
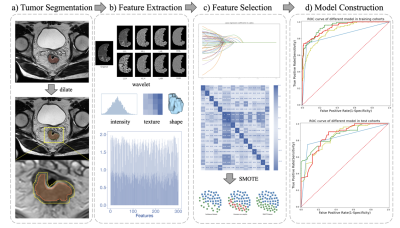
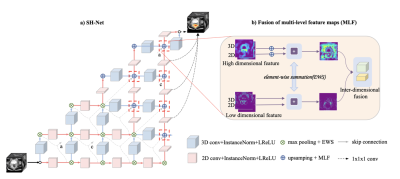
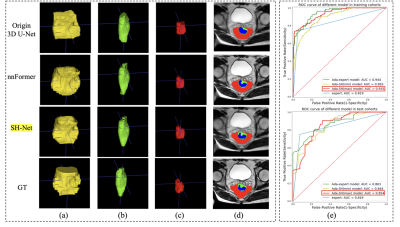
Figure 3. 3D segmentation results of the (a) circumferential margin, (b) the wall and the (c) tumor of the rectum in the test cohort. d) Results comparison in example slice of T2WI image (red: circumferential margin, green: the rectal wall, blue: tumor). e) ROC curve for different models in the training and test cohorts for EMVI diagnosis of rectal cancer.

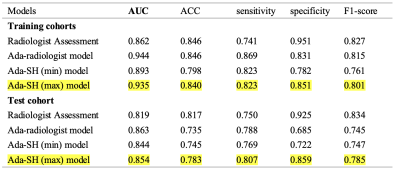
Table 2. Performance comparison of different models for EMVI prediction in the training and test cohorts. ACC: accuracy, Radiologist Assessment: T2WI-based diagnostic EMVI by radiologists, Ada-radiologist model: Adaboost Decision Tree radiomics model based on manually drawn ROIs by radiologists, Ada-SH model: spatial hybrid network (SH-Net) and Adaboost Decision Tree radiomics model, in which Ada-SH (min) means the minimum delineation of predicted tumor segmentation without dilation and Ada-SH (max) means maximum delineation of predicted tumor segmentation by dilation.