0241
Multicenter reproducibility of hand-crafted radiomics and deep-learning based features for biparametric prostate MRI1Department of Radiology, University of Turku, Raisio, Finland, 2Department of Radiology, Turku University Hospital, Turku, Finland, 3Department of Computing, University of Turku, Turku, Finland, 4Department of Urology, TYKS Turku University Hospital and University of Turku, Turku, Finland, 5Department of Pathology, TYKS Turku University Hospital, Turku, Finland, 6Department of Medical Physics, TYKS Turku University Hospital, Turku, Finland
Synopsis
Keywords: Radiomics, Cancer, Inter-site reproducibility, Deep Learning, bi-parametric MRI
In the current study, we aimed to explore reproducibility of various hand-crafted radiomics features, and deep learning autoencoder-based features within Gleason Grade Groups (GGG) using MULTI-IMPROD trial data. Differences between sites were evaluated with ANOVA test, corrected for GGG group, and multi-class AUC for GGG. We explored if systematic differences exist between the four centers taking part in the trial with conventionally used radiomic features. The results show differences between modalities, feature groups, and when intensity harmonization is applied for ADC.Introduction
Radiomic feature extraction and artificial intelligence (AI) in medical imaging has experienced significant progress over the past decade [1], and [2]. These methods are increasingly being used in prostate MRI [3]. However, reproducibility of various hand-crafted and deep learning based radiomic features for prostate MRI remain predominantly unknown. In a registered multicenter MULTI-IMPROD trial, a unique biparametric prostate MRI, IMPROD bpMRI [4,5] demonstrated potential to reduce the number of unmercenary biopsy procedures while improving detection of clinically significant prostate cancer. Prostate cancer aggressiveness was graded using Gleason Grade Groups (GGG) [6]. In the current study, we explored reproducibility of various hand-crafted radiomics features, and deep learning autoencoder-based features within GGG using MULTI-IMPROD trial data.Methods
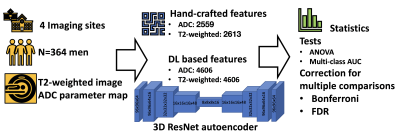
Between September 2014 and May 2017, 364 men with a clinical suspicion of PCa were prospectively enrolled at four different institutions in Finland (Turku, Pori, Tampere, Helsinki) into a prospective, registered validation trial (MULTI-IMPROD, NCT02241122) of IMPROD bpMRI in men with clinical suspicion of PCa. Enrolled men had two repeated PSA measurements ranging from 2.5-20.0 ng/mL and/or abnormal digital rectal exam (DRE).Datasets
IMPROD bpMRI was performed using body array coils (no endorectal coil) at 3 Tesla MRI (T) scanners in Turku (Verio, Siemens), Tampere (Skyra, Siemens), Helsinki (Skyra, Siemens), while a 1.5T (Aera, Siemens) MRI scanner was used in Pori. Imaging consisted of T2-weighted acquisitions in axial and sagittal planes, three separate diffusion weighted imaging (DWI) (5 b-values 0-500 s/mm2, 2 b-values 0-1500 s/mm2, 2 b-values 0-2000 s/mm2) and corresponding calculated apparent diffusion coefficient maps (ADCm) fitted using mono-exponential fit. In the current study, we used T2-weighted axial images (0.6×0.6×3.0mm3) together with Apparent Diffusion Coefficient (ADC) maps (2.0×2.0×3.0mm3) calculated from 5 b-values DWI, as biparametric images for feature extraction.
Image post-processing
The current study overview is shown in Figure 1. For each subject, ADC map was aligned to T2-weighted image. Whole gland and lesion (from 1-4 lesions per patient) segmentations were done by on voxel level by on radiologist with 10 years of experience in radiology separately on axial T2w and DWI (5 b-values 0-500 s/mm2) images. Radiomic feature extraction was performed using pyradiomics package [7], radiomics package for repeatable radiomics [8], and 3D autoencoder ResNet variant for prostate data, using 96x96x64 central region from image encompassing the whole gland. In total 2559 hand-crafted radiomic features were evaluated from ADC, 2613 for T2-weighted images. We also evaluated deep learning auto encoder based values, respectively.
Statistical Analysis
Differences between sites with ANOVA test (stats package 4.0.2), corrected for GGG group, were evaluate to explore if systematic differences exist between the four center taking part in the trial. All centers used the same IMPROD bpMRI protocol. Multi-class AUC (pROC package 1.16.2) was used to evaluate the classification potential for GGG. We compared overall inter-center reproducibility between features extracted from ADC and T2-weighted images, and reproducibility between radiomic feature set. For ADC, we evaluated effect of applying intensity normalization [9], where 64 subjects from site 1 were used to estimate intensity histogram parameters inside whole gland for normalization. We applied correction for multiple comparisons over number of evaluated measures (Bonferroni and FDR), so that corrected p-values<0.05 were considered statistically significant. All statistical evaluations were executed with R 4.0.2.
Results
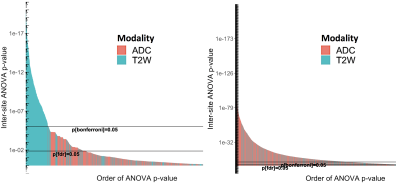
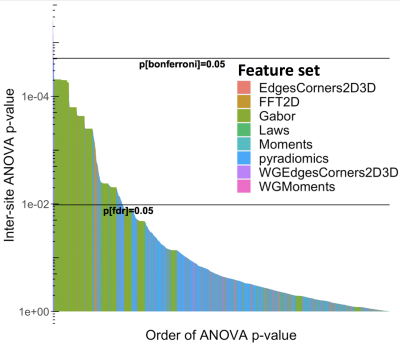
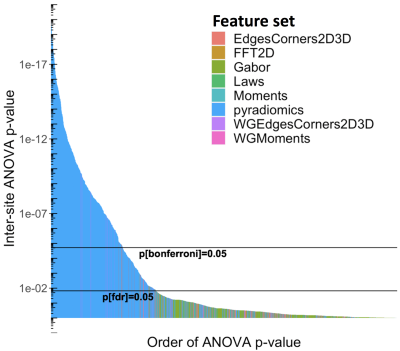
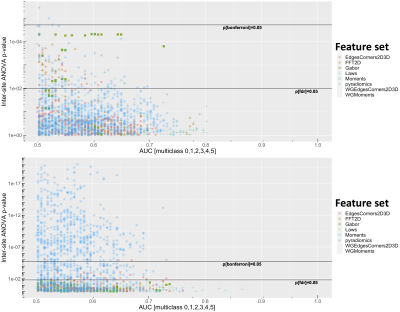
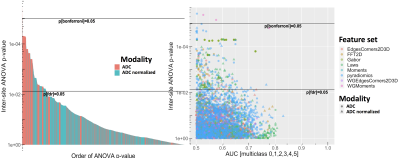
Differences between imaging sites, corrected for GGG, are shown in Figures 2-6, with statistically significant differences between sites with the features. Generally, more pronounced differences within set of hand-crafted radiomics were those derived from T2-weighted imaging, while no notable difference was found with deep learning extracted values (Figure 2). Within modality radiomics based on Gabor filter with ADC (Figure 3) were least robust against differences between sites, while with T2-weighted imaging (Figure 4) radiomics from pyradiomics package gave features demonstrating biggest inter-site differences. Further, when assessing for classification potential (Figure 5), best performance in terms of both general consistency between sites and AUC was found with individual features from the evaluated groups. Lastly, in evaluation of intensity harmonization before feature extraction, there was apparent benefit from using harmonization.Discussion
A number of radiomic features were found to be statistically significantly non-reproducible between sites. We speculate that T2-weighted image based radiomics had more poor inter-site performance due to higher spatial resolution, which in turn may affect the feature values to be more sensitive to differences between imaging sites. As intensity normalization had positive impact on reproducibility with ADC where the same acquisition sequence was used originally between sites, harmonization may be considered helpful to improve inter-site reproducibility. It is to be noted that most of the individual extracted features from T2-weighted images are not to be considered to depend on intensity values in absolute terms, while ADC derived features depend more on the intensity values because of lower spatial resolution. Based on our analysis, we consider reproducibility analysis to be potentially useful in informing deep learning architectures in designing and training.Conclusion
The results indicate that individual radiomic features individually or as part of machine learning or other applications aiming to generalize classification of prostate cancer should be used with care, as their numerical estimates may vary largely between imaging sites. Normalization of ADC between centers led to improved inter-site repeatability.Acknowledgements
HM was funded by Academy of Finland (#26080983).References
[1] Bera, K., Braman, N., Gupta, A., Velcheti, V. and Madabhushi, A., 2022. Predicting cancer outcomes with radiomics and artificial intelligence in radiology. Nature Reviews Clinical Oncology, 19(2), pp.132-146.
[2] Li, C., Li, W., Liu, C., Zheng, H., Cai, J. and Wang, S., 2022. Artificial intelligence in multiparametric magnetic resonance imaging: A review. Medical Physics.
[3] Michaely, H.J., Aringhieri, G., Cioni, D. and Neri, E., 2022. Current Value of Biparametric Prostate MRI with Machine-Learning or Deep-Learning in the Detection, Grading, and Characterization of Prostate Cancer: A Systematic Review. Diagnostics, 12(4), p.799.
[4] Jambor I, Boström PJ, Taimen P, et al. Novel biparametric MRI and targeted biopsy improves risk stratification in men with a clinical suspicion of prostate cancer (IMPROD Trial). J Magn Reson Imaging. 2017;46(4):1089–1095.
[5] Ettala, O., Jambor, I., Perez, I.M., Seppänen, M., Kaipia, A., Seikkula, H., Syvänen, K.T., Taimen, P., Verho, J., Steiner, A. and Saunavaara, J., 2022. Individualised non-contrast MRI-based risk estimation and shared decision-making in men with a suspicion of prostate cancer: protocol for multicentre randomised controlled trial (multi-IMPROD V. 2.0). BMJ open, 12(4), p.e053118.
[6] Loeb, S., Folkvaljon, Y., Robinson, D., Lissbrant, I.F., Egevad, L. and Stattin, P., 2016. Evaluation of the 2015 Gleason grade groups in a nationwide population-based cohort. European urology, 69(6), pp.1135-1141.Vancouver
[7] Van Griethuysen, J.J., Fedorov, A., Parmar, C., Hosny, A., Aucoin, N., Narayan, V., Beets-Tan, R.G., Fillion-Robin, J.C., Pieper, S. and Aerts, H.J., 2017. Computational radiomics system to decode the radiographic phenotype. Cancer research, 77(21), pp.e104-e107.
[8] Merisaari, H., Taimen, P., Shiradkar, R., Ettala, O., Pesola, M., Saunavaara, J., Boström, P.J., Madabhushi, A., Aronen, H.J. and Jambor, I., 2020. Repeatability of radiomics and machine learning for DWI: Short‐term repeatability study of 112 patients with prostate cancer. Magnetic resonance in medicine, 83(6), pp.2293-2309.
[9] Nyúl, L.G. and Udupa, J.K., 1999, June. New variants of a method of MRI scale normalization. In Biennial International Conference on Information Processing in Medical Imaging (pp. 490-495). Springer, Berlin, Heidelberg.
Figures