0239
The Influence of Variability and Uncertainty in the Clinical Reference on MRI Radiomics Modelling and Performance1Research Department, Hong Kong Sanatorium and Hospital, Hong Kong, Hong Kong, 2Department of Imaging and Interventional Radiology, The Chinese University of Hong Kong, Hong Kong, Hong Kong, 3Department of Diagnostics and Interventional Radiology, Hong Kong Sanatorium and Hospital, Hong Kong, Hong Kong
Synopsis
Keywords: Radiomics, Machine Learning/Artificial Intelligence, Data Modelling
Radiomics uses quantitative analysis of medical imaging based on machine learning techniques and has shown its potentials of aiding personalized clinical decisions. A high standard of clinical reference (or ground truth, endpoint) is vital in radiomics feature selection and modeling, but is commonly overlooked, and assumed to be perfect. However, in reality, there are uncertainties and variability in these clinical references due to many factors. We aim to quantitatively assess the influence of clinical reference uncertainty and variability on MRI Radiomics modeling via endpoint annotation permutation with different levels.Introduction
Radiomics utilizes quantitative analysis of medical imaging based on machine learning techniques and has potentials of aiding personalized clinical decisions on diagnosis and therapy [1], while its reliability and generalizability are still concerns and have yet to be further investigated [2].A high standard of clinical reference (or ground truth, endpoint) and its accurate annotation is vital in radiomics feature selection and modeling but is an overlooked factor of reliability in most radiomics studies. The clinical reference annotations are normally presumed to be perfect without bias/errors. However, in reality, there are uncertainties and variability in these clinical references due to either low-standard clinical reference (e.g. biopsy v.s. pathology), different criteria/definitions, or human-induced bias, disagreement, and errors in data collection/labeling [3]. In this study, we aim to quantitatively assess the influence of clinical reference uncertainty and variability on MRI Radiomics modeling via endpoint annotation permutation with different levels.
Methods
A publicly available radiomics data extracted from the PROSTATEx dataset [4-5] was used, which contains multi-parametric MRI of 260 prostate cancer (PC) patients (127 clinically significant (CS) PC and 133 non-clinically-significant PC (NCS-PC), based on the Gleason-score obtained by post-prostatectomy pathology). 265 IBSI-compliant radiomics features were extracted from T2W image (TSE, 0.5 x 0.5 x 3.6 mm3), DWI images (b-value=800 s/mm2, 2x2x3.6 mm3) and ADC maps. The data were separated into training (n=182) and testing datasets (n=78) with the CS-NCS ratio 2.3:1. Only the endpoints in the training dataset were perturbed by permutation with the levels of 5%, 10%, 50%, and 100%, and repeated 10 times, to simulate clinical reference uncertainties.Four different feature selection methods of Mann Whitney U test, Recursive Feature Elimination (RFE), Least Absolute Shrinkage and Selection Operator (LASSO), Minimum Redundancy Maximum Relevance (MRMR), and two classifiers of Random Forest classifiers, and LASSO were adopted to build seven radiomics models for each endpoint permutation. Area under the Curve (AUC) of ROC curve, sensitivity, specificity, and accuracy of each model were also calculated and compared using ANOVA test with Bonferroni correction.
Results
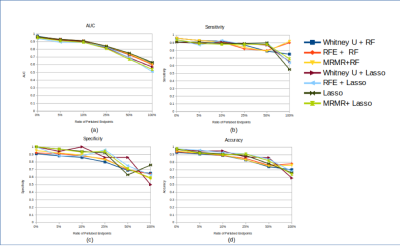
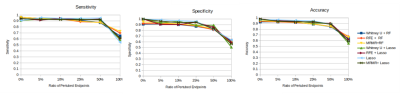
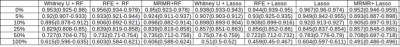
Permutation levels of 5%, 10%, 50%, and 100% resulted in 3.14% (2.20-3.85%), 5.71% (4.40-6.59%), 13.41% (10.44-16.48%), 26.26% (21.98-30.77%), and 49.12% (42.86-57.14%) of erroneous clinical references. A higher ratio of permutated endpoints resulted in more features selected except for Mann Whitney U test. T2_original_shape_Flatness and DWI_original_glrlm_ShortRunEmphasis are two features consistently selected using MRMR in all permutation levels. Four features (T2_original_shape_Flatness, T2_original_shape_LeastAxis, DWI_original_glrlm_RunLengthNonUniformity, and DWI_original_glrlm_ShortRunEmphasis) were consistently selected in 4 levels of permutation using RFE.Different combinations of feature selection and classification did not lead to significantly different model performance (p>0.05) for each permutation (Table 1). The mean AUCs with high permutation levels (25%: 0.830, 50%: 0.714, 100%: 0.583) were significantly lower (P<0.05) than those with low permutation levels (0%: 0.956, 5%: 0.917, 10%: 0.905) (fig.1 (a)) Sensitivity, specificity and accuracy also had the same decreasing trend in both training dataset with permutated endpoint and true endpoint, and testing dataset (figs. 1-3), without showing a significant difference for each permutation level. The mean accuracy of all models associated with higher permutation levels (50%: 0.793, 100%: 0.687) was significantly lower than that without permutation (0%: 0.96) (P<0.05).
Discussion
Uncertainties and variability of clinical reference endpoint could be caused by many factors, where some might be unavoidable. From the 7 models built using MRI Radiomics with permuted endpoints in different levels, the results showed that the performance of the models remained robust up until a certain level of permutation. All model performances were excellent up to the permutation levels of 25%, which corresponded to ~13% endpoint uncertainty, partly attributed to the high quality of the clinical reference of post-prostatectomy pathology and tumor segmentation by experts in the PROSTATEx dataset. The AUC decreased with the increasing level of permutation. For the permutation levels of 100%, the AUC was ~0.55, only slightly better (~5%) than random guessing (AUC=0.5), indicating that the developed radiomics models did reveal intrinsic tumor properties rather than overfit the permuted clinical references. Further, the models even trained with the permuted clinical references did not lead to significantly reduced performance in the testing set with the true references, also suggesting the robustness of radiomics modeling. Nevertheless, this study has some limitations in the retrospective and simulation study design and the utilization of only two classifiers in a single public dataset. Future studies with larger data on different diseases should be conducted for further validation.Conclusion
Our preliminary results showed that the low level (up to ~13%) of uncertainties and variabilities in clinical references might not significantly affect radiomics modeling. This study also suggested the importance of obtaining a high standard of clinical reference for radiomics studies.Acknowledgements
No acknowledgement found.References
1. Lambin P, Rios-Velazquez E, Leijenaar R, Carvalho S, van Stiphout RG, Granton P, Zegers CM, Gillies R, Boellard R, Dekker A, Aerts HJ. Radiomics: extracting more information from medical images using advanced feature analysis. Eur J Cancer 2012;48:441-6. 10.1016/j.ejca.2011.11.036
2. Xue C, Yuan J, Lo GG, Chang ATY, Poon DMC, Wong OL, Zhou Y, Chu WCW. Radiomics feature reliability assessed by intraclass correlation coefficient: a systematic review. Quant Imaging Med Surg. 2021 Oct;11(10):4431-4460. doi: 10.21037/qims-21-86. PMID: 34603997; PMCID: PMC8408801.
3. Novis DA, Zarbo RJ, Valenstein PA. Diagnostic uncertainty expressed in prostate needle biopsies. A College of American Pathologists Q-probes Study of 15,753 prostate needle biopsies in 332 institutions. Arch Pathol Lab Med. 1999 Aug;123(8):687-92. doi: 10.5858/1999-123-0687-DUEIPN. PMID: 10420224.
4. Song Y, Zhang J, Zhang Yd, Hou Y, Yan X, et al. (2020) FeAture Explorer (FAE): A tool for developing and comparing radiomics models. PLOS ONE 15(8): e0237587. https://doi.org/10.1371/journal.pone.0237587
5. Litjens G, Debats O, Barentsz J, Karssemeijer N, Huisman H. Computer-aided detection of prostate cancer in MRI. IEEE Trans Med Imaging. 2014;33(5):1083–92. Epub 2014/04/29. pmid:24770913.
Figures