0196
Deep learning improves breast IVIM estimation in better benign and malignant lesion differentiation
Shuhao Shi1, Lu Wang1, Jianfeng Bao2, Zhigang Wu3, Congbo Cai1, Zhong Chen1, Jiazheng Wang3, and Shuhui Cai1
1Department of Electronic Science, Xiamen University, Xiamen, China, 2Department of Radiology, First Affiliated Hospital of Zhengzhou University, Zhengzhou, China, 3MSC Clinical & Technical Solutions, Philips Healthcare, Shenzhen, China
1Department of Electronic Science, Xiamen University, Xiamen, China, 2Department of Radiology, First Affiliated Hospital of Zhengzhou University, Zhengzhou, China, 3MSC Clinical & Technical Solutions, Philips Healthcare, Shenzhen, China
Synopsis
Keywords: Breast, Cancer
Intravoxel incoherent motion (IVIM) with multiple b-values, as an advanced diffusion model, provides accurate identification of breast cancer. However, the IVIM-derived parameters vary greatly depending on different fitting methods, especially for parameters D* and f. In this study, we proposed a method for high-quality breast IVIM reconstruction based on deep neural network. Data analysis shows that our proposed method improves the visual quality of breast IVIM parametric maps with better benign and malignant breast lesion differentiation ability compared to the traditional least-square fitting method.Introduction
Breast cancer has become the most commonly diagnosed cancer in the world,1 and early diagnosis has a significant impact on breast cancer survival rates. Intravoxel incoherent motion (IVIM) magnetic resonance (MR) imaging is a noninvasive perfusion imaging technique that allows evaluation of tissue diffusion and microcapillary perfusion and has been proven to be valuable in the differential diagnosis of breast lesions.2 In practice, both the fitting method and the signal-to-noise ratio (SNR) of diffusion-weighted imaging (DWI) data have great influences on the estimated IVIM parametric maps,3,4 especially for breast DWI data which suffer a lot from low SNR. Accurate and precise IVIM bi-exponential quantification is still a mathematically challenging problem to be solved. It is generally acknowledged that deep neural networks have unmatched nonlinear fitting capability and have been effectively used to resolve challenging quantitative MR mapping problems.5,6 Here we introduce a deep neural network to estimate IVIM parameters from multi-b-value breast DWI data.Methods
MR imaging: MR images of 46 patients were retrospectively analyzed in the First Affiliated Hospital of Zhengzhou University from December 2015 to December 2017. A 3.0 T GE Discovery 750 superconducting MR scanner and an 8-channel breast-specific phase-controlled coil were used. Detailed imaging protocols were as follows: (1) DWI: b = 0, 20, 50, 100, 150, 200, 400, 800, 1200, 1600, 2000, 2500, 3000 and 4000 s/mm2, number of excitation corresponding to 1, 1, 1, 1, 1, 2, 2, 2, 4, 4, 6, 6, 8 and 10, TR/TE = 3600/76 ms, FOV = 320 mm × 320 mm, matrix = 128 × 192, layer thickness/interlayer spacing 4/1 mm. (2) Contrast-enhanced MRI: Axial-position volume imaging sequence Vibrant, TR/TE = 3.9/1.7 ms, FOV = 360 × 360 mm, matrix = 320 × 320, layer thickness/interlayer spacing = 1.4/1.0 mm.Data analysis: Figure 1 shows the process of IVIM parameters estimation based on deep learning. Synthetic data were employed for deep neural network training. To complete the nonlinear mapping, a 5-level U-Net with skip connections was introduced. The trained network model was tested on 46 patients. For comparison, the IVIM parameters were also estimated using a "segmented" least-square (LS) fitting method. 7 The region of interests (ROIs) were drawn on the grayscale DWI images with b = 1, 000 s/mm2. We extracted the mean, extreme, and heterogeneity indexes of ROIs with histogram analysis.
Statistical analysis: Statistical analyses were conducted using SPSS 25.0 and MedCalc 20.0.22. All continuous variables were subjected to the Shapiro-Wilk normality test and the Levene homogeneity of variance test. Comparison between groups was performed using t test or Mann-Whitney U test. Receiver operating characteristic (ROC) curve was used to evaluate the diagnostic value of histogram indices for benign and malignant breast lesion differentiation. The area under the curve (AUC) of each ROC was compared using the DeLong test. Significance was defined as p < 0.05.
Result
Figure 2 shows the IVIM parametric maps estimated from the two methods of a patient diagnosed with fibroadenoma. Lesion margins were significantly improved and noise was largely eliminated in the parametric maps estimated from the deep neural network compared to the segmented LS method. The time for IVIM parameter estimation is significantly reduced from approximately 44 s for the LS method to about 30 ms for the neural network.Table 1 and Table 2 show a comparison of histogram metrics derived from the two methods between benign and malignant breast lesions. In Table 1, among the parameters estimated from LS, D (minimum, mean, median), and f (minimum, mean) were significantly higher in malignant breast lesions than in benign lesions, whereas D (skewness, kurtosis), D* (skewness), and f (variance) were significantly lower in malignant breast lesions. In Table 2, among the parameters estimated from the deep neural network, D (minimum, mean, median), D* (skewness), and f (minimum) were significantly higher in malignant breast lesions than in benign lesions, while D (skewness, kurtosis), and D* (mean, median) were significantly lower in malignant breast lesions.
Figure 3 shows the ROC curves of the two methods for benign and malignant breast lesion differentiation. Among all the histogram metrics of estimated IVIM parameters, the D*-median has the largest AUC (0.833) and Youden index (0.625) for the proposed method and the D-median has the highest AUC (0.808) and Youden index (0.622) for the LS algorithm. The DeLong test shows no significant difference between the two ROCs (p = 0.7399).
Discussion and conclusion
This study proposes a method for breast IVIM parameter estimation based on deep learning. We solved the problem of large dataset required for deep neural network training by introducing synthetic data. Experimental findings demonstrate the feasibility of utilizing deep neural network for IVIM parameter estimation in place of the conventional LS method. IVIM parametric maps obtained from the proposed method have clearer texture structure and less background noise. The data analysis results show that the IVIM parameters obtained from our proposed method also improve the diagnosis of benign and malignant breast lesions. In summary, the proposed method enhances the visual quality of breast IVIM parametric maps, improves the benign and malignant breast lesion differentiation, and significantly reduces the parameter estimation time.Acknowledgements
This work was supported by the National Natural Science Foundation of China under grant numbers 11775184, 82071913 and 22161142024.
References
1. Sung H, Ferlay J, Siegel RL, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021; 71: 209-249.2. Ma W, Mao J, Wang T, et al. Distinguishing between benign and malignant breast lesions using diffusion weighted imaging and intravoxel incoherent motion: A systematic review and meta-analysis. Eur J. Radiol. 2021;141: 109809.
3. Vidic I, Jerome NP, Bathen TF, et al. Accuracy of breast cancer lesion classification using intravoxel incoherent motion diffusion-weighted imaging is improved by the inclusion of global or local prior knowledge with Bayesian methods. J. Magn. Reson. Imaging 2019; 50: 1478-1488.
4. Wittsack HJ, Lanzman RS, Mathys C, et al. Statistical evaluation of diffusion-weighted imaging of the human kidney. Magn. Reson. Med. 2010; 64: 616-622.
5. Cai CB, Wang C, Zeng YQ, et al. Single-shot T2 mapping using overlapping-echo detachment planar imaging and a deep convolutional neural network. Magn. Reson. Med. 2018; 80: 2202-2214.
6. Zhang J, Wu J, Chen SJ, et al. Robust single-shot T2 mapping via multiple overlapping-echo acquisition and deep neural network. IEEE Trans. Med. Imaging 2019; 38: 1801-1811.
7. Sigmund E, Cho G, Kim S, et al. Intravoxel incoherent motion imaging of tumor microenvironment in locally advanced breast cancer. Magn. Reson. Med. 2011; 65: 1437-1447.
Figures
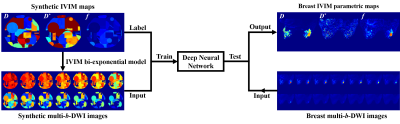
Figure 1. Framework of the estimation of IVIM derived parametric maps from DWI data by the deep neural network. Synthetic multi-b-DWI data: b = 0, 20, 50, 100, 150, 200, 400, 800, 1200, 1600, 2000, 2500, 3000 and 4000 s/mm2.
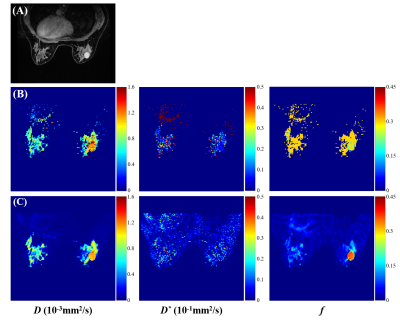
Figure 2. IVIM parametric maps of a patient with fibroadenoma. (A) Contrast-enhanced T1 weighted image. (B) IVIM parametric maps estimated from "segmented" least-square (LS) algorithm. (C) IVIM parametric maps estimated from the deep neural network.
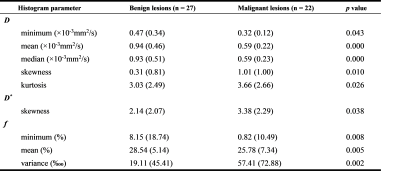
Table 1. Differences in IVIM histogram parameters estimated from the LS algorithm between benign and malignant breast lesions. Continuous variables with normal distribution were presented as mean (standard deviation) values; non-normal variables were presented as median (interquartile range) values.

Table 2. Differences in IVIM histogram parameters estimated from the deep neural network between benign and malignant breast lesions. Continuous variables with normal distribution were presented as mean (standard deviation) values; non-normal variables were presented as median (interquartile range) values.
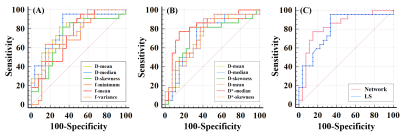
Figure 3. (A) ROC curves of IVIM parameters estimated from the LS algorithm. (B) ROC curves of IVIM parameters estimated from the deep neural network. (C) Comparison of optimal ROC curves (D* median for CNN, D median for LS) of the two methods (p = 0.7399).
DOI: https://doi.org/10.58530/2023/0196