0165
Federated Learning for Utilizing Multi-Institutional Prostate MRI with Diverse Histopathology
Abhejit Rajagopal1, Katya Redekop2, Anil Kemisetti1, Rishi Kulkarni3, Steven Raman3, Karthik Sarma3, Kirti Magudia4, Corey Arnold2,3, and Peder Larson1
1Radiology and Biomedical Imaging, UCSF, San Francisco, CA, United States, 2Electrical Engineering, UCLA, Los Angeles, CA, United States, 3Radiology, UCLA, Los Angeles, CA, United States, 4Radiology, Duke University, Durham, NC, United States
1Radiology and Biomedical Imaging, UCSF, San Francisco, CA, United States, 2Electrical Engineering, UCLA, Los Angeles, CA, United States, 3Radiology, UCLA, Los Angeles, CA, United States, 4Radiology, Duke University, Durham, NC, United States
Synopsis
Keywords: Machine Learning/Artificial Intelligence, Cancer, federated learning
Prostate cancer screening and diagnosis from MRI is extremely challenging, and current machine learning algorithms suffer in cross-institutional generalizability. Federated learning is a way to alleviate these issues by combining multi-center data without aggregating or homogenizing data. To enable this for prototype-stage algorithms, we introduce FLtools, a lightweight python library with re-usable federated learning components available freely at https://federated.ucsf.edu. We use this federated learning system to train a 3D UCNet on bi-parametric MRI and paired prostate biopsy data from two University of California hospitals, demonstrating dramatic improvements in cross-site generalization accuracy in clinically-significant lesion classification.Introduction
Early prostate cancer detection and staging from MRI is extremely challenging for both radiologists and deep learning algorithms [1], but the potential to learn from large and diverse datasets remains a promising avenue to increase their performance within and across institutions [2]. To enable this for prototype-stage algorithms, where the majority of existing research remains, we introduce a flexible federated learning framework for cross-site training, validation, and evaluation of custom deep learning prostate cancer detection algorithms.Specifically, we introduce an abstraction of prostate cancer groundtruth that represents diverse annotation and histopathology data. We maximize use of this groundtruth if and when they are available using UCNet, a custom 3D UNet that enables simultaneous supervision of pixel-wise, region-wise, and gland-wise classification. We leverage these modules to perform cross-site federated training using 1400+ heterogeneous multi-parameteric prostate MRI exams from two University of California hospitals.
Methods: Datasets and Histopathology Groundtruth
Bi-parametric magnetic resonance imagery (bp-MRI) prostate exams are composed of T2-weighted images, and apparent diffision coefficient (ADC) maps derived from and dynamic contrast-enhanced (DCE) MRI. bp-MRI provides complementary information on the content and restriction of water, but may vary in appearance due to subtle differences in choice of MR scanner hardware and physiologic variability in healthy prostate and tumors (Figure 1). In this work, we register bp-MRI to a consistent spatial resolution of [0.66,0.66,2.24]mm. The UCSF dataset consisted of 679 training, 96 validation, and 198 testing exams. The UCLA dataset consisted of 737 training, 101 validation, and 148 testing exams. A key difference between these prostate-MRI datasets is in the corresponding histopathology derived from prostate biopsy. Specifically, the UCSF exams include MRI-identified lesion segmentations and ISUP grade groups (derived from Gleason patterns) for both MRI-targeted lesions and systematic biopsy sites, which differ in the granularity of radiographic localization. The UCLA exams include lesion segmentations and only the maximum ISUP grade group of all lesion biopsies, and no systematic biopsy data.Methods: Federated Learning Toolkit
We introduce a design pattern for federated learning (FL) that separates model development and FL implementation code, providing a feature-rich FL development environment that is meant to expose essential functionality of local gradient computation and federated weight updating without requiring any re-implementation. Our design pattern is composed of 3 elements: a model abstraction, a data abstraction, and a model-agnostic federated toolkit.Our federated toolkit, FLtools, is a lightweight python library available freely at https://federated.ucsf.edu, which includes baseline implementations of various FL routines (structured to enable backend compatibility with Nvidia's NVFlare [3] and Flower [4]), but which are crucially reusable across models and FL experiments (Figure 2). Specifically, FLComponents defines an abstraction and interface for FL training, aggregation, and serialization algorithms, enabling reuse of federated components across projects.
The data abstraction is composed of two parts. For the general FL design pattern, we utilize user-built dataloaders so FL code can work independent of the learning task. For the specific prostate-MRI classification problem, the dataloader yields bp-MRI $$$x\in\mathbb{R}^{3 \times X \times Y \times Z}$$$ and binary region masks $$$y\in\mathbb{R}^{R\times X\times Y\times Z}$$$ representing the $$$R$$$ regions with histopathology data. Crucially, we encode the histopathology data using the matrix $$$z\in \mathbb{Z}^{R\times 2}$$$, representing a supervision signal {0,1,2} and a maximum ISUP grade group (0-negative, 1-5) for each region. The supervision signal is used to dynamically select learning objectives applicable to the type of histopathology groundtruth on a region-by-region basis, as described next.
Methods: UCNet Architecture
We define the UCNet architecture using a 3D UNet backbone with a fully-connected classification output head, taking MRI as input and predicting 3D lesion segmentation maps, ISUP grade group maps, and region-wise histograms representing clinically-significant cancer (Figure 3). UCNet is designed to handle diverse groundtruth histopathology data available for prostate cancer via the dynamically-populated multi-task training objective:$$ \mathcal{L}(x, y, y_\text{gg}, z)=\beta_1\mathcal{L}_\text{region-classifier}+\beta_2\mathcal{L}_\text{GGmap-hist}+\beta_3\mathcal{L}_\text{GGmap}+\beta_4\mathcal{L}_\text{segmentation}
$$
where $$$\bar{\beta}\in{Z}_{[0,1]}^4$$$ is populated based on the availability of groundtruth data and the desired network function, $$$\mathcal{L}_\text{segmentation}$$$ is a standard Dice-cross-entropy loss, $$$\mathcal{L}_\text{region-classifier}$$$ and $$$\mathcal{L}_\text{GGmap}$$$ are standard categorical cross-entropy losses, and $$$\mathcal{L}_\text{GGmap-hist} = \mathcal{L}_\text{hist-strong} + \mathcal{L}_\text{hist-high}$$$ is a novel distribution based loss defined by the uncertainty in biopsy data groundtruth. That is, for MRI-identified lesions with an expected homogeneous cancer profile, we define:
$$
\mathcal{L}_\text{hist-strong}(z,y_\text{gg},\hat{h}) =\frac{1}{|R^\alpha|}\sum_{r \in R^\alpha}\sum_{k=1}^K y_\text{gg}[r,k]\log{\hat{h}[r,k]}
$$
whereas for systematic biopsy areas with an expected inhomogeneous or homogenous cancer profile, we define:
$$
\mathcal{L}_\text{hist-high}(z,y_\text{gg},\hat{h})=\frac{1}{|R^\beta|}\sum_{r\in R^\beta}\quad\sum_{{k \; >\underset{k}{\mathrm{argmax}} \; y_\text{gg}[r]}}^K y_\text{gg}[r,k] \log{\hat{h}[r,k]}
$$
where $$$R$$$ is an encoding of the histopathology signal, and $$$K=2$$$ for binary classification.The net effect of these losses is to suppress the proportion of voxels representing grade groups not supported by the histopathology data.
Results and Discussion
Table 1 depicts the comparative performance on the radiographic lesion classification task, demonstrating a dramatic 9.5-14.8% improvement in cross-site test-set accuracy and specificity using federated learning. That is, UCSF and UCLA local models performed well on data from their own sites (Figure 4,Row2), but did not generalize well across sites until using federated learning (Figure 4, Rows3-4).Conclusion
Our FL system and UCNet model were able to handle highly heterogeneous prostate MRI, patient distributions, and histopathology groundtruth without needing to transfer, pool, or homogenize data at a single location. The clinical impact of federated CS-PCa detection models is improved generalization accuracy and physician confidence in deployed models.Acknowledgements
This work was supported by NIH/NIBIB grant #F32EB030411, NIH/NCI grants #R01CA229354 and #R21CA220352, a Society for Abdominal Radiology Morton A. Bosniak Research Award, a RSNA Research Resident/Fellow Grant, the Cancer League and Helen Diller Family Comprehensive Cancer Center at UCSF.References
[1] Westphalen, Antonio C., et al. "Variability of the positive predictive value of PI-RADS for prostate MRI across 26 centers: experience of the society of abdominal radiology prostate cancer disease-focused panel." Radiology 296.1 (2020): 76-84.
[2] Kairouz, Peter, et al. "Advances and open problems in federated learning." Foundations and Trends® in Machine Learning 14.1–2 (2021): 1-210.
[3] NVIDIA Corporation, "Nvidia federated learning application runtime environment", https://github.com/NVIDIA/NVFlare, 2021.
[4] Beutel, Daniel J., et al. "Flower: A friendly federated learning research framework." arXiv preprint arXiv:2007.14390 (2020).
Figures
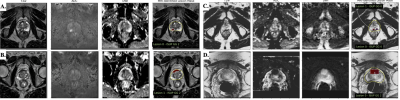
Figure 1: Intra- and inter-site variations of multiparametric MRI data. (A,B) Natural variation between appearance of ISUP grade group 2 lesions (countoured) in UCLA data. (C,D) Large variation in the apparent MR contrast and size of lesion annotations (bounding boxes) in UCSF data.
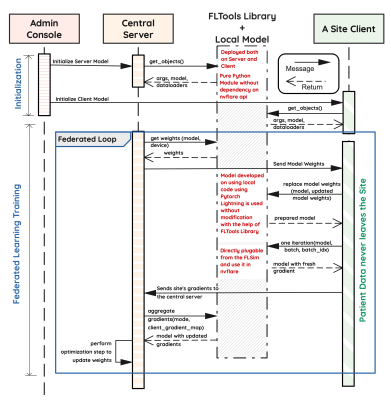
Figure 2: Modular federated system architecture depicting the FLtools v0.1 abstraction and data exchanges between the centralized server and each client.
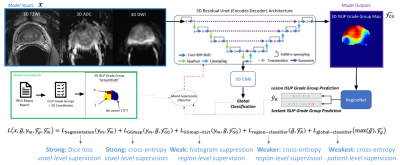
Figure 3: UCNet Architecture, depicted here with a 3D residual UNet backbone, histopathology-based histogram suppression, and regional classification modules. In this paper, UCNet takes registered 3D mp-MRI as input and produces as output: lesion segmentation maps, 1-hot-encoded cancer grading maps (for classification of clinically-significant prostate cancer, $$$K=2$$$), and per-region classifications ($$$\mathcal{L}_\text{global}$$$ not trained).
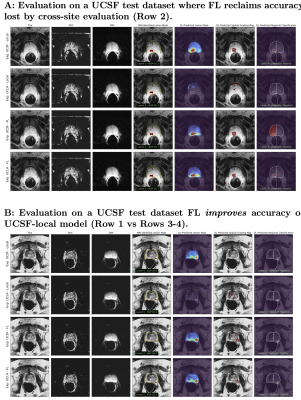
Figure 4: Evaluation of UCNet on UCSF dataset. (A) depicts a transverse slice of an exam with a MRI-identified ISUP GG 2 lesion where the UCLA-local model performs poorly, but both federated models (Rows 3-4) achieves the same level of accuracy as UCSF-local model. (B) depicts a transverse slice of an exam where both local models perform poorly, but federated models correctly classify this lesion as CS-PCa. Checkpoint chosen by UCLA stopping criteria (Row 3) outperforms checkpoint chosen by UCSF (Row 4), highlighting that neither site has sufficient data to generalize well on their own.
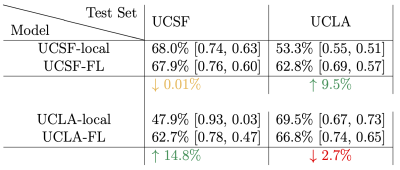
Table 1: Region-wise Lesion Binary Classification Accuracy. Bracketed numbers indicate true negative rate (TNR, specificity) and true positive rate (TPR, sensitivity), respectively. Results indicate dramatic improvements in the cross-site generalization accuracy of the UCNet model when trained via federated learning (without data aggregation or homogenization).
DOI: https://doi.org/10.58530/2023/0165