0162
A transformer-based framework for liver stiffness classification using multi-modality body MRI in children and adults1Department of Radiology, Cincinnati children's hospital medical center, Cincinnati, OH, United States, 2Department of Radiology, Cincinnati children's hospital medical center, 45429, OH, United States, 3University of Wisconsin-Madison, Madison, WI, United States, 4Michigan Medicine, University of Michigan, Ann Arbor, MI, United States, 5New York University Langone Health, New York, NY, United States, 6Cincinnati children's hospital medical center, Cincinnati, OH, United States
Synopsis
Keywords: Machine Learning/Artificial Intelligence, Liver
Magnetic resonance elastography (MRE) provides a noninvasive method to quantify liver stiffening, a surrogate biomarker for monitoring liver fibrosis. However, the availability of MRE remains limited, especially outside the United States, in part due to cost. This study aims to develop a deep learning-based approach for stratifying liver stiffness using multiparametric MRI images from pediatric and adult patients from multiple sites. We performed multi-site ten-fold cross-validation and achieved an AUROC of 0.80 for liver stiffness stratification. These results demonstrate that our proposed deep learning model may provide a means for categorical estimation of liver stiffening without dedicated elastography.Introduction
Despite advances in chronic liver disease (CLD) management, CLD remains a leading cause of death in both children and adults in the United States and worldwide [1, 2]. CLD can potentially lead to advanced liver fibrosis in some patients, which increases liver tissue stiffness [3]. End-stage liver fibrosis is associated with considerable morbidity and mortality, and may require liver transplantation as definitive treatment. Magnetic resonance elastography (MRE) is well established to measure liver stiffness, a noninvasive quantitative surrogate marker for liver fibrosis [4]. MRE evaluates tissue stiffness by placing a vibrating passive driver over the right upper quadrant of the abdomen that generates shear waves in the liver [1, 4, 5]. Shear waves are imaged using a modified phase contrast pulse sequence, with shear wave speed increasing with increased liver stiffness. Although MRE technology may reduce the demand for liver biopsy in some patients, it has associated drawbacks including additional exam time, patient discomfort, and added healthcare costs related to additional imaging, post-processing, hardware, and software. In the current project, we propose deep learning algorithms that can be used to determine liver stiffness from readily available conventional MRI anatomic images.Methods
Study cohortIn this HIPAA-compliant, IRB-approved multi-site retrospective study, clinical MRI images acquired between 2011 and 2020 were retrieved from the clinical Picture Archiving and Communicating Systems (PACS) of four institutions, including Cincinnati Children's Hospital Medical Center (CCHMC), New York University (NYU), University of Michigan (UM), and the University of Wisconsin (UW). We collected 2,846 axial T1-weighted gradient echo fat-saturated and T2-weighted fast spin-echo fat-saturated MRI sequences from patients with known or suspected chronic liver disease. For each patient, MRE liver stiffness measurements were extracted from electronic health records to serve as the reference standard for stratifying liver stiffness. Patients were categorized into two groups (no/mild liver stiffening [<3 kPa] vs. moderate/severe liver stiffening [≥3 kPa]) [1]. Demographic and liver stiffness characteristics of our patient population are shown in Table 1 by site.
Deep learning model
Our deep learning model consists of an input imaging layer and two blocks, including transfer learning and adaptive learning blocks. First, the input imaging layer has two distinct input channels for T1- and T2-weighted images, respectively (Figure 1). The input is a stack (S=11) of axial 2D T1-weighted gradient echo fat-saturated and T2-weighted fast spin-echo fat-saturated MR images with a size of 224×224. Second, we designed a transfer learning block by reusing the weights of the state-of-art Swin Transformer [6] that was trained based on ~14.2 million natural images. Swin Transformer builds a hierarchical representation by merging small-sized patches and gradually integrating neighboring patches in deeper layers to achieve scale-invariance, as shown in Figure 1 (c). The hierarchical architecture has low computational complexity regarding image size due to the computation of self-attention locally. Swin Transformer splits an input image into non-overlapping patches using a shifting window approach, as shown in Figure 1 (d). Finally, we employ an adaptive learning block with trainable layers to learn the individual latent features from 11 images of the liver from each subject. The adaptive learning block contains global average pooling (GAP), batch normalization (BN), and two fully connected (FC) layers with [512, 256] neurons and a two-way softmax classifier to classify the severity of liver stiffness.
Due to the imbalanced subject ratio (e.g., <3 vs. ≥3 kPa = ~2:1 in the current study), a weighted cross-entropy loss function was used. The diagnostic performance of our model was assessed using receiver operating characteristic (ROC), area under the curve (AUC), sensitivity, specificity, and accuracy. We applied a stratified multi-site ten-fold cross-validation (Figure 2), where subjects from four study sites were proportionally split into training, validation, and testing data while preserving the ratio of samples for each site and severity groups.
Results
In multi-site ten-fold cross-validation, our proposed deep learning model was able to categorically classify the severity of liver stiffening using combined T1- and T2-weighted images with an accuracy [mean ± standard deviation] of 74.1 ± 0.02, specificity of 73.7 ± 0.04, sensitivity of 71.3 ± 0.04 and AUROC of 0.80 ± 0.01. Further results using single modality such as T1 or T2 weighted images are listed in Table 2.Conclusions
The proposed deep learning model demonstrated moderate stratification performance on a large, multi-site combined pediatric and adult dataset for categorically classifying the severity of liver stiffening using anatomic T1- and T2-weighted MRI data. Further improvements in performance can be achieved by incorporating additional pulse sequences as well as clinical data, and ultimately may lead to fewer MRE imaging and/or percutaneous liver biopsy procedures.Acknowledgements
This work was supported by the National Institutes of Health [R01-EB030582, R01-EB029944, R01-NS094200, and R01-NS096037]; Academic and Research Committee (ARC) Awards of Cincinnati Children's Hospital Medical Center. The funders played no role in the design, analysis, or presentation of the findings.References
[1] Li, H., He, L., Dudley, J. A., Maloney, T. C., Somasundaram, E., Brady, S. L., ... & Dillman, J. R. (2021). DeepLiverNet: a deep transfer learning model for classifying liver stiffness using clinical and T2-weighted magnetic resonance imaging data in children and young adults. Pediatric radiology, 51(3), 392-402. [2] Lee, Jeong Hyun, et al. "Deep learning with ultrasonography: automated classification of liver fibrosis using a deep convolutional neural network." European radiology 30.2 (2020): 1264-1273. [3] Trout, A. T., Anupindi, S. A., Gee, M. S., Khanna, G., Xanthakos, S. A., Serai, S. D., & Dillman, J. R. (2020). Normal Liver Stiffness Measured with MR Elastography in Children. Radiology, 297(3), 663-669. [4] He, L., Li, H., Dudley, J. A., Maloney, T. C., Brady, S. L., Somasundaram, E., ... & Dillman, J. R. (2019). Machine learning prediction of liver stiffness using clinical and T2-weighted MRI radiomic data. American Journal of Roentgenology, 213(3), 592-601. [5] Pollack, B. L., Batmanghelich, K., Cai, S. S., Gordon, E., Wallace, S., Catania, R., & Borhani, A. A. (2021). Deep learning prediction of voxel-level liver stiffness in patients with nonalcoholic fatty liver disease. Radiology: Artificial Intelligence, 3(6). [6] Liu, Z., Lin, Y., Cao, Y., Hu, H., Wei, Y., Zhang, Z., & Guo, B. (2021). Swin transformer: Hierarchical vision transformer using shifted windows. In Proceedings of the IEEE/CVF International Conference on Computer Vision (pp. 10012-10022).Figures
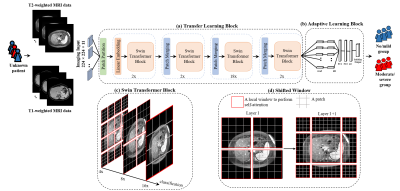
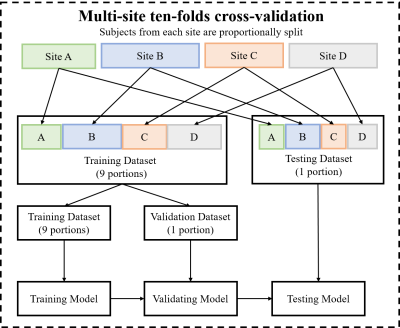
Figure 2 The ten-fold cross-validation experiments flow chart used to validate the model is presented above. Multi-site cross-validation splits the cohorts from four study sites into training, validation, and testing sets while preserving the ratio of samples for each site and medical diagnosis.
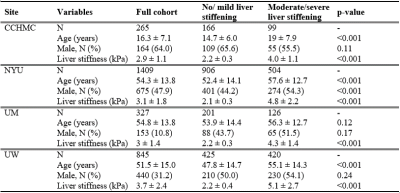
Table 1 Demographic and liver stiffness characteristics of patients for four sites, including Cincinnati Children's Hospital Medical Center (CCHMC), New York University (NYU), University of Michigan (UM), and University of Wisconsin (UW).
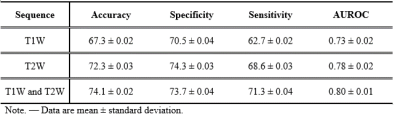
Table 2 Diagnostic performance of the deep learning model following multi-site ten-fold cross-validation for categorically classifying patients, using anatomic axial T1 and T2 weighted MR images.