0157
Reducing annotation burden in MR segmentation: A novel contrastive learning loss with multi-contrast constraints on local representations1Department of Electrical and Computer Engineering, University of Arizona, Tucson, AZ, United States, 2Department of Medical Imaging, University of Arizona, Tucson, AZ, United States, 3College of Medicine, University of Arizona, Tucson, AZ, United States, 4Department of Radiology, Houston Methodist Hospital, Houston, TX, United States, 5Program in Applied Mathematics, University of Arizona, Tucson, AZ, United States, 6Department of Biomedical Engineering, University of Arizona, Tucson, AZ, United States
Synopsis
Keywords: Machine Learning/Artificial Intelligence, Machine Learning/Artificial Intelligence, Representational Learning
The availability of limited labeled data motivates the use of self-supervised pretraining techniques for deep learning (DL) models. Here, we propose a novel contrastive loss that pushes/pulls local representations within an image based on representational constraints from co-registered multi-contrast MR images that share similar underlying parameters. For multi-organ segmentation tasks in T2-weighted images, pretraining a DL model using the proposed loss function with constraints from co-registered echo images from a radial TSE acquisition, can help reduce annotation burden by 60%. On two independent datasets, proposed pretraining improved Dice scores compared to random initialization and pretraining with conventional contrastive loss.Introduction
Contrastive learning, a form of representation learning, has shown promising results in pretraining deep learning (DL) models to mitigate the lack of labeled data in MR image segmentation tasks1,2. Contrastive loss3 optimizes weights θ of the DL model $$$\Psi_\theta(.)$$$ such that the distance between representations $$$(\Psi_\theta(x_a),\Psi_\theta(x_b))$$$ of pairs of inputs $$$(x_a,x_b)$$$ approximate their semantic similarity.In this work, we propose a novel contrastive loss pre-training framework that uses representational constraints from multi-contrast MR images to guide the contrastive loss in learning discriminating local representations within an image. Local regions in multi-contrast MR images with similar underlying tissue types (e.g., local regions within liver in abdomen) share similar signal characteristics and as such, should produce similar feature representations in the representational space. We demonstrate the applicability of the proposed pretraining framework to a) reduce annotation burden and b) improve segmentation performance in downstream multi-organ segmentation tasks in two different MR datasets.
Methods: Contrastive Loss with multi-contrast constraints
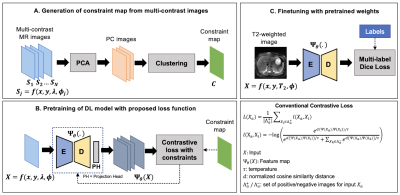
An illustration of the proposed framework is presented in Figure-1. Let $$$\Psi_\theta(X)$$$ be the feature representation for an image $$$X=f(x,y,\lambda,\phi)$$$ obtained using a forward pass through the DL model. Here, $$$\lambda$$$ and $$$\phi$$$ denote underlying tissue-specific parameter(s) (T1, T2, Ktrans, etc.) and other imaging parameters, respectively.For a randomly selected p×p local region $$$m_a$$$ in $$$\Psi_\theta(X)$$$, and its positive ($$$Λ_a^+$$$) and negative ($$$Λ_a^-$$$) similarity sets, the contrastive loss $$$L(m_a)$$$ is given by
$$L(m_a )= \frac{1}{|Λ_a^+ |}∑_{m_i∈Λ_a^+}l(m_a,m_i) $$
$$l(m_a,m_i)=-\log(\frac{\exp^{d(\Psi(m_a ),\Psi(m_i ))/\tau}}{\exp^{d(\Psi(m_a ),\Psi(m_i ))/\tau}+∑_{x_k∈Λ_a^-}\exp^{d(Ψ(m_a ),Ψ(m_k ))/\tau}})$$
Previous works in classification1 and image segmentation4,5 have explored augmentation strategies to generate sets of regions/images $$$Λ_a^+$$$ and $$$Λ_a^-$$$ whose representations should be brought closer to/pushed away from those of $$$m_a$$$. We propose to identify these regions using a) a representational constraint map C generated from multi-contrast MR images that share similar underlying parameters and b) hard negative mining in the representational space6,7. We first identify $$$c_a$$$, a set of regions that share the same class (signal characteristics) as $$$m_a$$$ in the corresponding constraint map followed by regions $$$b_a$$$, whose representations are most similar to $$$m_a$$$ in the current training iteration (e.g., Top-k neighbors using a cosine similarity metric). We propose to impose the following constraints on the local representations during contrastive learning:
$$Λ_a^+=\{m_i: m_i \in c_a⋂ b_a\}$$
$$Λ_a^- =\{m_i: m_i \in \bar{c}_a⋂ b_a\}$$
Methods: Pre-training the Deep Learning Model
Consider a set of N multi-contrast MR images $$$S=\{S_1,S_2,…,S_N,; S_j=f(x,y,\lambda,\phi_j )\}$$$ co-registered with image $$$X=f(x,y,\lambda,\phi)$$$. We generate a representational constraint map (Figure-1A) by first performing principal component analysis on $$$S$$$ to retain $$$M<N$$$ principal components (PC) for dimensionality reduction and denoising, followed by applying an unsupervised clustering algorithm (K-Means, clusters K) on the retained PC images.We use a 2D-encoder-decoder architecture as our DL model and the pretraining is performed in multiple stages. First, the DL encoder is pretrained to embed global semantic information1,4. A warm start is provided to improve clustering of representations by pretraining with conventional contrastive loss4 for 10 epochs. The model is then trained using the proposed contrastive loss with multi-contrast constraints for 150 epochs.
Methods: Downstream Segmentation Task
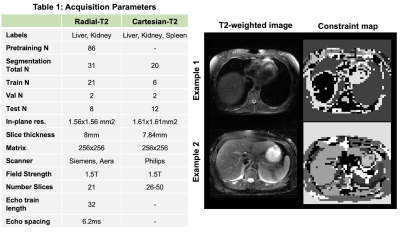
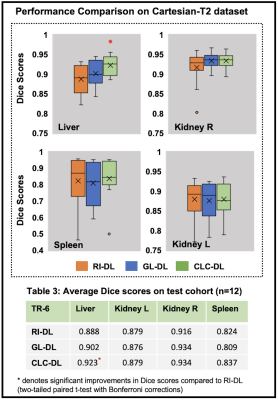
We demonstrate the utility of pretraining DL model using the proposed loss for multi-organ segmentation tasks in two different abdomen T2-weighted datasets: a) Radial Turbo Spin Echo (TSE) dataset (Radial-T2, n=117) with co-registered composite T2-weighted and echo images (TEs) from a single k-space acquisition8 and b) a public Cartesian TSE dataset9,10,11 (Cartesian-T2, n=20) (Figure-2A). The Radial-T2 images were bias corrected and de-streaked using CACTUS12. All T2-weighted images were intensity normalized using a histogram-based contrast stretching. A subset of Radial-T2 (n=31) labeled for a previous liver segmentation study13 were used here again for finetuning.We train three different DL models with weights initialized using: a) random initialization (RI-DL), b) global and local contrastive loss4 (GL-DL), and c) proposed contrastive loss with multi-contrast constraints (CLC-DL). Both GL-DL and CLC-DL were pretrained once on the remaining (n=86) Radial-T2 images and finetuned on the two segmentation datasets. The constraint maps (Figure-2) for CLC-DL were generated from co-registered echo images (N=32, M=3, K=30), encoding the underlying T2 parameter of the tissues.
Results and Discussion
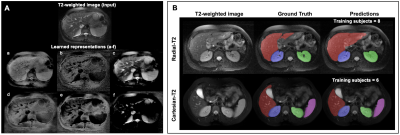
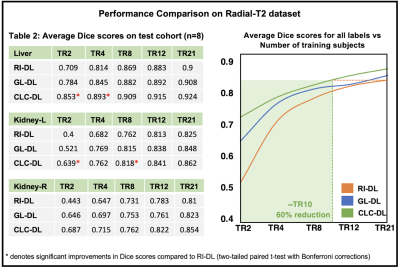
Figure-3A gives an insight into representations a DL model learns for a given T2-weighted image. On the segmentation tasks in Radial-T2 images, the average Dice scores of all DL models (Figure-4) improve with increasing number of training subjects. With multi-contrast constraints, CLC-DL trained only on 8 subjects performs comparable to RI-DL trained on all labeled data (n=21), leading to a 60% reduction in annotation effort. When using all labeled data for training, CLC-DL outperforms RI-DL and GL-DL models by 2% and 3-5% in Dice scores for liver and kidneys, respectively. Similar trends were observed in Cartesian-T2 where CLC-DL, pretrained on Radial-T2 images, yielded significant Dice score improvements in liver segmentation task when all models were trained with only 6 subjects (Figure-5). CLC-DL's performance on Cartesian-T2 also attests to its generalizability.Conclusion
We observed that pretraining a DL model using the proposed self-supervised contrastive loss with multi-contrast constraints on the representational space reduced the annotation burden for the downstream segmentation task, by as much as 60% compared to random initialization. When all available labeled data is used, the proposed framework yielded further improvements in terms of Dice scores compared to random initialization or conventional contrastive loss.Acknowledgements
We would like to acknowledge grant support from the National Institutes of Health (CA245920 and EB031894) and the Technology and Research Initiative Fund (TRIF) Improving Health Initiative.References
[1] Chen T, Kornblith S, Norouzi M, Hinton G. A Simple Framework for Contrastive Learning of Visual Representations. PMLR; 2020. https://github.com/google-research/simclr. Accessed January 18, 2021.
[2] Bai, W, Chen, C, Tarroni, G, Duan, J, Guitton, F, Petersen, SE, Guo, Y, Matthews, PM, Rueckert, D Self-supervised learning for cardiac mr image segmentation by anatomical position prediction. In: International Conference on Medical Image
[3] Hadsell R, Chopra S, LeCun Y. Dimensionality reduction by learning an invariant mapping. In: Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition. Vol 2.; 2006. doi:10.1109/CVPR.2006.100
[4] Chaitanya K, Erdil E, Karani N, Konukoglu E. Contrastive learning of global and local features for medical image segmentation with limited annotations. arXiv Prepr arXiv200610511. 2020
[5] Umapathy L, Fu Z, Philip R, Martin D, Altbach M, Bilgin A. (2022). Learning to segment with limited annotations: Self-supervised pretraining with regression and contrastive loss in MRI. arXiv preprint arXiv:2205.13109.
[6] Zhuang C, Zhai A, Yamins D. Local Aggregation for Unsupervised Learning of Visual Embeddings. 2019. IEEE/CVF International Conference on Computer Vision (ICCV), 6001-6011.
[7] F Wang and H Liu. Understanding the Behaviour of Contrastive Loss. 2021. IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), 2021, pp. 2495-2504, doi: 10.1109/CVPR46437.2021.00252.
[8] Altbach MI, Outwater EK, Trouard TP, Krupinski EA, Theilmann RJ, Stopeck AT, Kono M, Gmitro AF. Radial fast spin-echo method for T2-weighted imaging and T2 mapping of the liver. J Magn Reson Imaging. 2002 Aug;16(2):179-89. doi: 10.1002/jmri.10142. PMID: 12203766.
[9] A.E. Kavur, N.S. Gezer, M. Barış, S. Aslan, P.-H. Conze, et al. "CHAOS Challenge - combined (CT-MR) Healthy Abdominal Organ Segmentation", Medical Image Analysis, Volume 69, 2021. https://doi.org/10.1016/j.media.2020.10195010
[10] A.E. Kavur, M. A. Selver, O. Dicle, M. Barış, N.S. Gezer. CHAOS - Combined (CT-MR) Healthy Abdominal Organ Segmentation Challenge Data (Version v1.03) [Data set]. Apr. 2019. Zenodo. http://doi.org/10.5281/zenodo.3362844
[11] A.E. Kavur, N.S. Gezer, M. Barış, Y.Şahin, S. Özkan, B. Baydar, et al. Comparison of semi-automatic and deep learning-based automatic methods for liver segmentation in living liver transplant donors, Diagnostic and Interventional Radiology, vol. 26, pp. 11–21, Jan. 2020. https://doi.org/10.5152/dir.2019.19025
[12] Fu Z, Johnson K, Altbach MI, Bilgin A. Cancellation of streak artifacts in radial abdominal imaging using interference null space projection. Magn Reson Med. 2022; 88( 3): 1355- 1369. doi:10.1002/mrm.29285
[13] Umapathy L, Keerthivasan MB, Galons JP, et al. A comparison of deep learning convolution neural networks for liver segmentation in radial turbo spin echo images. In: Proceedings of International Society for Magnetic Resonance in Medicine.; 2018:2729. https://arxiv.org/abs/2004.05731.
Figures