0156
COLADA: Contrastive Learning for highly accelerated MR Image Reconstruction
Mevan Ekanayake1,2, Zhifeng Chen1,3, Kamlesh Pawar1, Gary Egan1,4, Mehrtash Harandi2, and Zhaolin Chen1,3
1Monash Biomedical Imaging, Monash University, Melbourne, Australia, 2Department of Electrical and Computer Systems Engineering, Monash University, Melbourne, Australia, 3Department of Data Science and AI, Monash University, Melbourne, Australia, 4School of Psychological Sciences, Monash University, Melbourne, Australia
1Monash Biomedical Imaging, Monash University, Melbourne, Australia, 2Department of Electrical and Computer Systems Engineering, Monash University, Melbourne, Australia, 3Department of Data Science and AI, Monash University, Melbourne, Australia, 4School of Psychological Sciences, Monash University, Melbourne, Australia
Synopsis
Keywords: Machine Learning/Artificial Intelligence, Image Reconstruction, self-supervised learning, contrastive learning, accelerated reconstruction, fastMRI
Most deep learning methods for MR image reconstruction heavily depend on supervised learning on fully sampled reference data, hence, lack generalizability on out-of-distribution inputs such as higher accelerations. To reduce the models’ dependency on fully sampled reference data and to leverage large cohorts of undersampled MR measurements, we propose a self-supervised framework that extracts contrastive features between different accelerations of a given scan and the rest of the scans in the dataset, which we then utilize for the downstream reconstruction task. Our quantitative and qualitative analysis demonstrates the superiority of the proposed framework for highly accelerated MR image reconstruction.Introduction
Deep learning (DL) methods have demonstrated a dramatic improvement in MR image reconstruction performance1-3. The success of these DL methods often depends on supervised training using large datasets with fully sampled MR measurements. However, this direct dependency on fully sampled references has made the modern DL methods lack generalizability to out-of-distribution data, abnormalities in anatomical structures, adversarial conditions such as noise and varied higher acceleration factors4. One approach to improve models’ generalizability is to train a single model on several different datasets. However, in medical imaging applications, it is difficult to obtain fully sampled large datasets due to physiological and physical constraints which also highlights the importance of exploring self-supervised methods5. To this end, we propose Contrastive Learning for highly accelerated MR Image Reconstruction (COLADA) based on self-supervised contrastive learning. In particular, we learn a novel contrastive MR feature space that can effectively be utilized for the downstream reconstruction task. Our proposed framework shows superior performance, especially at exceedingly high accelerations, compared to state-of-the-art DL models.Methods
Our contrastive learning approach randomly undersamples MR images to generate a range of accelerations (e.g., 4X, 6X, 8X, and 10X) for a single MRI dataset and optimizes a contrastive learning model among those generated images. The idea here is to capture the common high-level features from a particular dataset with various generative sampling schemes and discriminate undesired features from other datasets to improve model robustness. In other words, by enforcing a contrastive model, we intend to pull the latent representations of different accelerations of a given dataset together and push latent representations of other datasets away as seen in Fig. 1. Our model consists of two main components: (i) Self-supervised Contrastive Learning model and (ii) Reconstruction model.Self-supervised Contrastive Learning Model
As seen in Fig. 2(a), we utilized a single channel U-Net6 as the contrastive feature extractor, $$$f(⋅)$$$ to extract contrastive representations and an MLP Mixer7 as the non-linear head, $$$g(⋅)$$$ to reduce the learned contrastive representations to feature vectors suitable for the loss computation. We utilized the Normalized Temperature-scaled Cross-Entropy loss8 (NT-Xnet) to train the network by considering the different accelerations of the same scan as positive samples and the rest of the scans in the minibatch as negative samples. Thus, the loss function for a positive pair of examples $$$(x_i,x_j)$$$ is as below:
$$\ell_{i,j}=\frac{\text{exp}(\text{sim}(z_i,z_j)/ \tau)}{\sum_{k=1}^{pN}\Gamma_{ik}⋅\text{exp}(\text{sim}(z_i,z_k)/ \tau)}$$
where $$$z=g(f(x))$$$, $$$sim(u,v)=\frac{u^T v}{‖u‖‖v‖}$$$, and $$$\Gamma_{ik}=\begin{cases} 1 & \text{if } k \neq i,\\ 0 & \text{otherwise}\\ \end{cases}
$$$. $$$\tau$$$ is a temperature scaling factor which we set at $$$0.07$$$. In our experiments, we generated 4X, 6X, 8X, and 10X accelerations of a single scan, thus $$$p=4$$$ and the minibatch size, $$$N=16$$$. We utilized random 1D sampling masks9 to undersample the k-space data.
Reconstruction Model
After training the Contrastive feature extractor on a large cohort of undersampled data, we discarded the non-linear head and freeze the trainable parameters of the Contrastive feature extractor. Then we connected a cascaded reconstruction network which consists of Dual-channels U-Nets and Data consistency blocks as seen in Fig. 2(b) and trained the cascaded network on various accelerations (4X, 6X, 8X, and 10X) using $$$\ell_1$$$ loss while keeping the weights of the contrastive feature extractor fixed. We found out that implementing k-space correction and enforcing the contrastive features to learn the missing k-space information performs well.
Dataset
We sampled 6306 and 1586 transverse brain MRI scans from the fastMRI brain dataset9 (which were obtained on 3 and 1.5 Tesla MR scanners) for training and validation, respectively. The dataset contained raw k-space data belonging to four imaging sequences (T1, T2, T1POST, and FLAIR).
Experiments and Results
We compared the reconstruction performance of COLADA with that of two state-of-the-art benchmark models, i.e., the fastMRI baseline9 (U-net) and the Cascaded Network with data consistency10 (D5C5). All models were trained using 4X, 6X, 8X, and 10X accelerations and all imaging sequences. We evaluated the reconstruction performance of the models by generating 4X, 10X, and 16X accelerations for each scan in the validation set and assessing with respect to NMSE, PSNR, and SSIM. Note that none of the models were trained using 16X datasets.The quantitative scores in Fig. 3 show the superiority of the proposed method. We observe that, especially, at extremely higher accelerations such as 16X, conventional DL methods fail drastically whereas our method has a relatively small degradation in performance. The same observation could be seen in Fig. 4 where the U-Net produces blurry outputs and the D5C5 catastrophically fail to reconstruct 16X accelerations whereas our proposed framework performs commendably while preserving fine features of the MR images.
Discussion and Conclusions
Experimental results show that the learned contrastive features can be effectively used to reconstruct highly accelerated MR images. The concept of optimizing feature space using self-supervised contrastive learning will subsequently facilitate training on a large cohort of data without fully sampled references which could benefit the feature extraction as well as the downstream reconstruction by leveraging a large amount of publicly available datasets. To the best of our knowledge, this is the first attempt at utilizing self-supervised contrastive feature learning across different accelerations for MRI reconstruction with promising results, especially for very high acceleration factors.Acknowledgements
The authors are grateful for support from the Australian Research Council Linkage grant LP170100494 and the Australian Research Council Discovery grant DP210101863.References
1. Sriram et al., ‘End-to-End Variational Networks for Accelerated MRI Reconstruction’, in Medical Image Computing and Computer Assisted Intervention – MICCAI 2020, Cham, 2020, pp. 64–73.2. B. Zhu, J. Z. Liu, S. F. Cauley, B. R. Rosen, and M. S. Rosen, ‘Image reconstruction by domain-transform manifold learning’, Nature, vol. 555, no. 7697, Art. no. 7697, Mar. 2018.
3. M. Ekanayake, K. Pawar, M. Harandi, G. Egan, and Z. Chen, ‘Multi-head Cascaded Swin Transformers with Attention to k-space Sampling Pattern for Accelerated MRI Reconstruction’. arXiv, Jul. 18, 2022.
4. M. J. Muckley et al., ‘Results of the 2020 fastMRI Challenge for Machine Learning MR Image Reconstruction’, IEEE Trans. Med. Imaging, vol. 40, no. 9, pp. 2306–2317, Sep. 2021.
5. B. Yaman, S. A. H. Hosseini, S. Moeller, J. Ellermann, K. Uğurbil, and M. Akçakaya, ‘Self-supervised learning of physics-guided reconstruction neural networks without fully sampled reference data’, Magnetic Resonance in Medicine, vol. 84, no. 6, pp. 3172–3191, 2020.
6. O. Ronneberger, P. Fischer, and T. Brox, ‘U-Net: Convolutional Networks for Biomedical Image Segmentation’, in Medical Image Computing and Computer-Assisted Intervention – MICCAI 2015, vol. 9351, N. Navab, J. Hornegger, W. M. Wells, and A. F. Frangi, Eds. Cham: Springer International Publishing, 2015, pp. 234–241.
7. I. O. Tolstikhin et al., ‘MLP-Mixer: An all-MLP Architecture for Vision’, in Advances in Neural Information Processing Systems, 2021, vol. 34, pp. 24261–24272.
8. T. Chen, S. Kornblith, M. Norouzi, and G. Hinton, ‘A Simple Framework for Contrastive Learning of Visual Representations’, in Proceedings of the 37th International Conference on Machine Learning, Nov. 2020, pp. 1597–1607.
9. J. Zbontar et al., ‘fastMRI: An Open Dataset and Benchmarks for Accelerated MRI’, arXiv:1811.08839 [physics, stat], Dec. 2019.
10. J. Schlemper, J. Caballero, J. V. Hajnal, A. N. Price, and D. Rueckert, ‘A Deep Cascade of Convolutional Neural Networks for Dynamic MR Image Reconstruction’, IEEE Trans. Med. Imaging, vol. 37, no. 2, pp. 491–503, Feb. 2018.
Figures
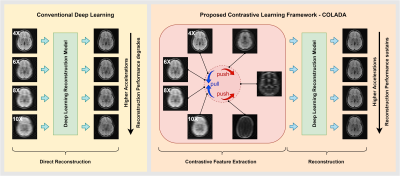
Figure 1: The overview of the proposed Contrastive Learning Framework - COLADA and how it differs from conventional deep learning
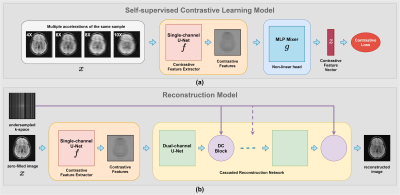
Figure 2: The
two main components of the proposed Contrastive Learning Framework - COLADA (a)
Self-supervised
Contrastive Learning model (b) Reconstruction
model
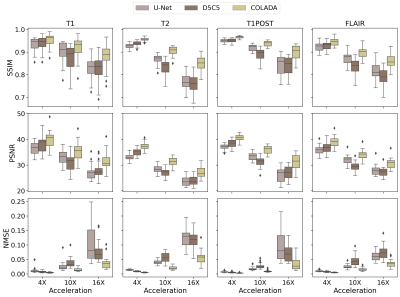
Figure 3: Quantitative
Reconstruction performance comparison of T1, T2, T1POST, and FLAIR images under
4X, 10X, and 16X accelerations with respect to Normalized Mean Square Error (NMSE), Peak
Signal-to-Noise Ratio (PSNR), and Structural Similarity (SSIM)
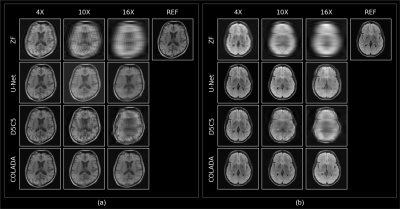
Figure 4: Qualitative
Reconstruction performance comparison under 4X, 10X, and 16X accelerations of a
(a) T1 scan (b) FLAIR scan
DOI: https://doi.org/10.58530/2023/0156