0088
Radiomic nomograms from pre-treatment magnetic resonance imaging-based may predict high-risk cytogenetic status in multiple myeloma1Second Hospital of Lanzhou University, Lanzhou, China
Synopsis
Keywords: Skeletal, Skeletal
Our study aimed to develop and validate one or more clinically relevant radiomic nomograms based on multisequence MRI radiomic features to predict the pre-treatment HRC status of patients with MM. We developed radiomic nomograms for 14 models at a single center using clinically obtained whole-spine MRI images of 159 patients with MM. This study revealed that radiomic features of pre-treatment MRI images are associated with HRC status in patients with MM. Among the proposed models, the nomograms of the FT2, FT2+1, and FT2+2+1 models were identified as outstanding at distinguishing patients with HRC from those with non-HRC status.
Abstract
Purpose: Multiple myeloma (MM) is a highly heterogeneous disease caused by a series of genetic and epigenetic events. It is the second most common hematologic malignancy after lymphoma and is associated with approximately 100,000 deaths annually worldwide (1). Cytogenic abnormalities (CA) are observed in almost all patients with MM (2). CA is an independent risk factor affecting the prognosis of MM (3). In addition, it has been shown that clonal heterogeneity and clonal evolution tend to be pronounced in these patients (4). Second, CA status can be used to assess response to therapy (5), the RVd regimen (lenalidomide/bortezomib/dexamethasone) is the backbone of MM chemotherapy (6); it has been associated with complete remission rates in up to 50% of patients. However, patients with HRC are usually resistant to chemotherapy with the RVd regimen (7). In contrast, induction, consolidation, and maintenance therapy using autologous hematopoietic stem cell transplantation combined with KRd regimens (carfilzomib, lenalidomide, dexamethasone) can improve survival rates up to 72% (8). Therefore, CA status is crucial for the selection of a MM treatment regimen and assessment of treatment outcome. The diagnosis of CA relies on the collection of tissue specimens and molecular biology tests such as fluorescence in situ hybridization(FISH)(9, 10). However, obtaining tissue specimens for CA testing usually requires bone marrow aspiration biopsy, which is invasive and may cause complications such as bleeding and infection in patients with hematologic disorders. Other limitations of this method include inadequate or inappropriate tissue sampling due to tumor spatial heterogeneity. In addition, the process of histological specimen analysis is cumbersome, complex, and expensive. Therefore, novel non-invasive methods of CA status determination are required. Radiomics is a high-throughput extraction and computational analysis method used to obtain potentially valuable high-dimensional information about tumor heterogeneity from medical images in a manner that is superior to that of the human eye (11). The value of radiomic features as imaging predictors for cancer diagnosis, treatment response, and prognosis, including in MM, has been demonstrated (12). Two previous studies have suggested the feasibility of using whole-spine MRI-based radiomics to predict high-risk cytogenetic (HRC) status (13, 14). However, these two studies were small (n=50 or 89) and failed to construct clinically applicable nomograms. Consequently, further studies are required in this field. This study aimed to develop and validate one or more clinically relevant radiomic nomograms based on multisequence MRI radiomic features to predict the pre-treatment HRC status of patients with MM.Materials and methods: Patients with MM (71 and 88 HRC and non-HRC, respectively) identified by fluorescence in situ hybridization were randomly divided into training (n=111) and test (n=48) sets. The regions of interest(ROIs) was assessed by T1-weighted imaging (T1WI), T2-weighted imaging (T2WI) and fat-suppressed T2-weighted imaging (FS-T2WI) in the sagittal position. The ROIs were determined and manually labeled by three readers. 14 models(combined radiomics models: T1, T2, FT2, T1+2, FT2+1, FT2+2, and FT2+2+1 models; radiomics models: T1-age, T2-age, FT2-age, T1+2-age, FT2+1-age, FT2+2-age, and FT2+2+1-age models) using linear logistic regression were developed using T1WI、T2WI and FS-T2WI to stratify of CA in MM. Nomogram performance was evaluated and compared using C-index,bootstrapping, accuracy, sensitivity, specificity, positive predictive value, negative predictive value, and Akaike information criterion.
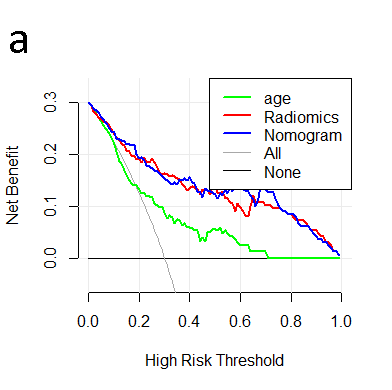
Results: The FT2 single-sequence model, FT2+1 double-sequence model, and FT2+2+1 multisequence model radiomics nomograms excelled at predicting HRC; the training set C-index of 0.80, 0.84 and 0.88, the test set C-index of 0.80, 0.84 and 0.84, respectively. These improvements to the C-indexes were validated using the 1000-times bootstrapping method; Favorable clinical application was observed using decision curve analysis.
Conclusions: The FT2 single-sequence, FT2+1 double-sequence, and FT2+2+1 multisequence models performed well at differentiating HRC and non-HRC states of MM. We present a non-invasive radiomics approach to determine the HRC status of patients with MM prior to treatment using clinically acquired multiparametric MRI images. The results of this study may aid clinical decision-making, including individualized treatment planning for MM.
Acknowledgements
Suwei LiuReferences
1. Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2021. CA Cancer J Clin 2021;71:7-33 2. Soekojo CY, Chung TH, Furqan MS, Chng WJ. Genomic characterization of functional high-risk multiple myeloma patients. Blood Cancer J 2022;12:24
3. Hose D, Beck S, Salwender H, Emde M, Bertsch U, Kunz C et al. Prospective target assessment and multimodal prediction of survival for personalized and risk-adapted treatment strategies in multiple myeloma in the gmmg-mm5 multicenter trial. J Hematol Oncol 2019;12:65
4. Neuse CJ, Lomas OC, Schliemann C, Shen YJ, Manier S, Bustoros M et al. Genome instability in multiple myeloma. Leukemia 2020;34:2887-2897
5. Sonneveld P, Avet-Loiseau H, Lonial S, Usmani S, Siegel D, Anderson KC et al. Treatment of multiple myeloma with high-risk cytogenetics: A consensus of the international myeloma working group. Blood 2016;127:2955-2962
6. Cowan AJ, Green DJ, Kwok M, Lee S, Coffey DG, Holmberg LA et al. Diagnosis and management of multiple myeloma: A review. Jama 2022;327:464-477
7. Morgan GJ, Davies FE, Gregory WM, Bell SE, Szubert AJ, Navarro Coy N et al. Cyclophosphamide, thalidomide, and dexamethasone as induction therapy for newly diagnosed multiple myeloma patients destined for autologous stem-cell transplantation: Mrc myeloma ix randomized trial results. Haematologica 2012;97:442-450
8. Kumar SK, Jacobus SJ, Cohen AD, Weiss M, Callander N, Singh AK et al. Carfilzomib or bortezomib in combination with lenalidomide and dexamethasone for patients with newly diagnosed multiple myeloma without intention for immediate autologous stem-cell transplantation (endurance): A multicentre, open-label, phase 3, randomised, controlled trial. Lancet Oncol 2020;21:1317-1330
9. Smith SC, Althof PA, Dave BJ, Sanmann JN. High-risk cytogenetics in multiple myeloma: Further scrutiny of deletions within the igh gene region enhances risk stratification. Genes Chromosomes Cancer 2020;59:569-574
10. Callander NS, Baljevic M, Adekola K, Anderson LD, Campagnaro E, Castillo JJ et al. Nccn guidelines® insights: Multiple myeloma, version 3.2022. J Natl Compr Canc Netw 2022;20:8-19
11. Gardin I, Grégoire V, Gibon D, Kirisli H, Pasquier D, Thariat J et al. Radiomics: Principles and radiotherapy applications. Crit Rev Oncol Hematol 2019;138:44-50
12. Guiot J, Vaidyanathan A, Deprez L, Zerka F, Danthine D, Frix AN et al. A review in radiomics: Making personalized medicine a reality via routine imaging. Med Res Rev 2022;42:426-440
13. Liu J, Zeng P, Guo W, Wang C, Geng Y, Lang N et al. Prediction of high-risk cytogenetic status in multiple myeloma based on magnetic resonance imaging: Utility of radiomics and comparison of machine learning methods. J Magn Reson Imaging 2021;54:1303-1311
14. Liu J, Wang C, Guo W, Zeng P, Liu Y, Lang N et al. A preliminary study using spinal mri-based radiomics to predict high-risk cytogenetic abnormalities in multiple myeloma. Radiol Med 2021;126:1226-1235
Figures
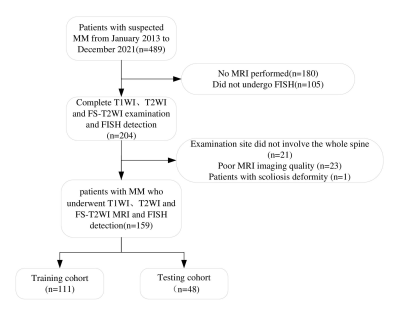
Flowchart of the patient selection process.
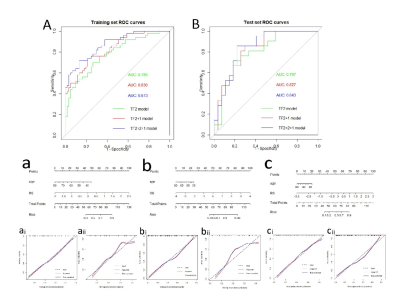
FT2, FT2+1, FT2+2+1 models nomogram training set, test set Receiver Operating Characteristic(A,B); FT2 single-sequence model nomogram and training set, test set calibration curves (a, ai, aii). FT2+1 double-sequence model nomogram and training set, test set calibration curves (b, bi, bii); FT2+2+1 multisequence model nomogram and training set, test set calibration curves (c, ci, cii).