0082
A Motion-Robust Slice-to-Volume Reconstruction Framework for Fetal Brain MRI1Massachusetts Institute of Technology, Cambridge, MA, United States, 2Computer Science, Vanderbilt University, Nashville, TN, United States, 3Fetal-Neonatal Neuroimaging and Developmental Science Center, Boston Children’s Hospital, Boston, MA, United States, 4Harvard Medical School, Boston, MA, United States, 5Computer Science and Artificial Intelligence Laboratory, Massachusetts Institute of Technology, Cambridge, MA, United States, 6Centre for Medical Image Computing, Department of Medical Physics and Biomedical Engineering, University College London, London, United Kingdom, 7Athinoula A. Martinos Center for Biomedical Imaging, Cambridge, MA, United States
Synopsis
Keywords: Image Reconstruction, Machine Learning/Artificial Intelligence
Volumetric reconstruction of fetal brains from multiple stacks of MR slices is challenging due to severe subject motion and image artifacts. We propose a deep learning method to solve the slice-to-volume reconstruction problem in two stages. First, a Transformer network is used to correct motion between slices by registering the input slices to a 3D canonical space. Second, an implicit neural network reconstructs the 3D volume by learning a continuous 3D representation of the fetal brain from the 2D observations. Results show that our method achieves high reconstruction quality and outperforms existing state-of-the-art methods in presence of severe fetal motion.Introduction
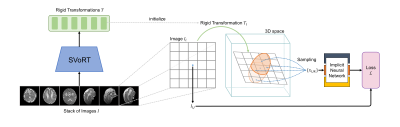
Reconstructing 3D volumes from motion-corrupted stacks of slices has shown promise in fetal brain MRI[1–3]. Existing slice-to-volume reconstruction (SVR) methods[3–5] estimate the slice transformations and the volume by performing slice-to-volume registration and super-resolution reconstruction alternatingly. However, this framework is vulnerable to severe subject motion and is time-consuming, especially when a high-resolution volume is desired. In this work, we propose a novel SVR framework that is robust to severe fetal motion and reconstructs high-resolution volumes efficiently. Our framework consists of two stages: 1) slice-to-volume registration by mapping stacks of slices to a 3D canonical space using a Transformer-based network, and 2) super-resolution reconstruction with implicit neural representation (INR).Methods
Traditional slice-to-volume registration methods[3] have a limited capture range of subject motion since the transformations are optimized by maximizing the local slice-to-volume consistency and therefore tend to get stuck in local minima. To address this problem, we use the Slice-to-Volume Registration Transformer (SVoRT)[6] to map stacks of slices to a 3D canonical space. The Transformer architecture[7] jointly processes the stacks of slices as a sequence and registers each slice by utilizing the global context from other slices, resulting in lower registration errors. The motion-corrected slices are then used to initialize the 3D reconstruction.Existing methods reconstruct 3D volumes by solving the following optimization problem,$$\min_{V,T}\sum_{i,j}\left( I_{ij} - \sum_k M_{ijk}V_k \right)^2,$$where $$$I_{ij}$$$ is the $$$j$$$-th pixel of the $$$i$$$-th acquired slice, $$$V_k$$$ is the $$$k$$$-th voxel of the volume, and $$$M_{ijk}$$$ is the coefficient in the forward acquisition model representing the slice transformation $$$T_i$$$, point spread function (PSF) and downsampling. This formulation bears some disadvantages. First, the discrete representations of volumes and PSFs might introduce discretization errors. Second, the algorithm reconstructs a volume only at a specific resolution and needs to be re-run when a different resolution is desired.
Recently, Neural Slice-to-Volume Reconstruction (NeSVoR)[8] was proposed for SVR in MRI, which learns a continuous representation of the fetal brain using INR[9–11]. NeSVoR adopts the following continuous forward model,$$\mu_{ij}=\int_\Omega M_{ij}(x)V_\theta(x)\text{d}x,\quad \sigma^2_{ij}=\int_\Omega M^2_{ij}(x)\beta_{i,\theta}(x)\text{d}x,$$where $$$\mu_{ij}$$$ and $$$\sigma^2_{ij}$$$ are the mean and variance of the pixel in the slices simulated by the forward model, $$$V_\theta(x)$$$ and $$$\beta_{i,\theta}(x)$$$ are continuous functions of spatial coordinates $$$x$$$, i.e., neural networks, which encode the intensity and uncertainty of the volume respectively. $$$M_{ij}(x)$$$ is the continuous PSF. Assuming a Gaussian model, NeSVoR solves the following optimization problem for the parameters in the networks and slice transformations,$$\min_{\theta,T}\frac{(\mu_{ij}-I_{ij})^2}{2\sigma_{ij}^2}+\frac{1}{2}\log\sigma_{ij}^2.$$After fitting the networks, NeSVoR is able to sample 3D volumes efficiently at arbitrary resolutions from the INR.
We adopted as a baseline three state-of-the-art SVR methods: SVRTK[3,12], SVR-GPU[4,13], and NiftyMIC[5,14]. For these baseline methods, we used the default hyperparameters in their open-sourced implementations. Experiments were performed on a system with an Intel Xeon 6238R CPU and an NVIDIA Tesla V100 GPU.
A fetal MRI dataset was collected to evaluate different SVR methods, which consisted of 484 T2-weighted MRI scans from fetuses with GA from 21 to 32 weeks. The data were acquired with in-plane resolution of 1-1.3mm, slice thickness of 2-4mm, TE=100-120ms, TR=1.4-1.8s. Each scan had three to ten stacks of slices acquired at different orientations. For preprocessing, the N4 bias field correction[15] was performed for each stack, and then brains are segmented from the slices using a segmentation network[5,16].
Results and Discussion
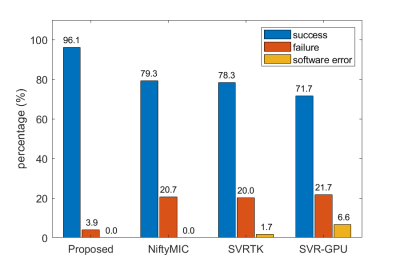
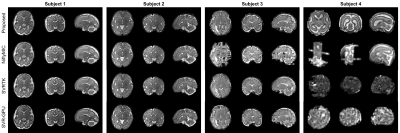
To evaluate the reconstruction quality, we classified the reconstructed volumes into three categories, success, failure (severe slice misalignment), and software error (no volume is generated). Fig. 2 summarizes the results. The success rates of all the baseline methods are less than 80%, while the proposed method achieves the highest success rate of 96%, demonstrating a significant improvement in robustness.Fig. 3 shows the reconstruction results of four fetal subjects. The reconstruction quality of the proposed method is similar to that of the baselines when there is little motion. However, the proposed method is more robust to severe subject motion and artifacts (subjects 3 and 4).
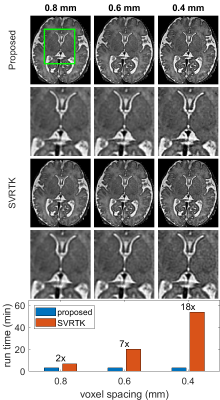
NeSVoR[8] learns a continuous representation of the fetal brain from which volumes at different resolutions can be sampled. In comparison, conventional methods reconstruct a volume by optimizing a discretized 3D array at a specific resolution. Therefore, the proposed method is more efficient when a high-resolution volume is desired. Fig. 4. shows the reconstructed volumes with different voxel spacings. The run time of SVRTK increases drastically as the voxel spacing decreases. In contrast, the run time of the proposed method is independent of the voxel spacing.
Fig. 5 illustrates the convergence of the INR reconstruction. The model is able to learn a high-quality representation of the underlying 3D volume from motion-corrected 2D slices in about one minute. The fast convergence of the proposed method demonstrates the potential of motion-robust online reconstruction of fetal MRI during scans.
Conclusion
In this work, we propose a novel method for SVR in fetal MRI. First, we adopt the SVoRT model to correct fetal motion by registering 2D slices to a 3D space, leading to a better-conditioned reconstruction problem. Second, the super-resolution reconstruction problem is solved efficiently with the implicit neural representation. Results on in utero data show that the proposed method achieves a higher success rate in reconstruction and a larger capture range of motion compared to other state-of-the-art methods.Acknowledgements
This research was supported by NIH U01HD087211, NIH R01EB01733, NIH HD100009, NIH NIBIB NAC P41EB015902, NIH 1R01AG070988-01, 1RF1MH123195-01 (BRAIN Initiative), ERC Starting Grant 677697, Alzheimer's Research UK ARUK-IRG2019A-003.References
1. Rousseau F, Glenn OA, Iordanova B, Rodriguez-Carranza C, Vigneron DB, Barkovich JA, et al. Registration-based approach for reconstruction of high-resolution in utero fetal MR brain images. Acad Radiol. 2006;13: 1072–1081.
2. Gholipour A, Estroff JA, Warfield SK. Robust super-resolution volume reconstruction from slice acquisitions: application to fetal brain MRI. IEEE Trans Med Imaging. 2010;29: 1739–1758.
3. Kuklisova-Murgasova M, Quaghebeur G, Rutherford MA, Hajnal JV, Schnabel JA. Reconstruction of fetal brain MRI with intensity matching and complete outlier removal. Medical Image Analysis. 2012. pp. 1550–1564. doi:10.1016/j.media.2012.07.004
4. Kainz B, Steinberger M, Wein W, Kuklisova-Murgasova M, Malamateniou C, Keraudren K, et al. Fast Volume Reconstruction From Motion Corrupted Stacks of 2D Slices. IEEE Trans Med Imaging. 2015;34: 1901–1913.
5. Ebner M, Wang G, Li W, Aertsen M, Patel PA, Aughwane R, et al. An automated framework for localization, segmentation and super-resolution reconstruction of fetal brain MRI. Neuroimage. 2020;206: 116324.
6. Xu J, Moyer D, Ellen Grant P, Golland P, Iglesias JE, Adalsteinsson E. SVoRT: Iterative Transformer for Slice-to-Volume Registration in Fetal Brain MRI. Lecture Notes in Computer Science. 2022. pp. 3–13. doi:10.1007/978-3-031-16446-0_1
7. Vaswani A, Shazeer N, Parmar N, Uszkoreit J, Jones L, Gomez AN, et al. Attention is all you need. Adv Neural Inf Process Syst. 2017;30. Available: https://proceedings.neurips.cc/paper/7181-attention-is-all-you-need
8. Xu J, Moyer D, Gagoski B, Iglesias JE, Ellen Grant P, Golland P, et al. NeSVoR: Implicit Neural Representation for Slice-to-Volume Reconstruction in MRI. doi:10.36227/techrxiv.21398868.v1
9. Mildenhall B, Srinivasan PP, Tancik M, Barron JT, Ramamoorthi R, Ng R. NeRF: representing scenes as neural radiance fields for view synthesis. Commun ACM. 2021;65: 99–106.
10. Martin-Brualla R, Radwan N, Sajjadi MSM, Barron JT, Dosovitskiy A, Duckworth D. NeRF in the wild: Neural Radiance Fields for unconstrained photo collections. arXiv [cs.CV]. 2020. pp. 7210–7219. Available: http://openaccess.thecvf.com/content/CVPR2021/html/Martin-Brualla_NeRF_in_the_Wild_Neural_Radiance_Fields_for_Unconstrained_Photo_CVPR_2021_paper.html
11. Sitzmann V, Martel JNP, Bergman AW, Lindell DB, Wetzstein G. Implicit neural representations with periodic activation functions. arXiv [cs.CV]. 2020. pp. 7462–7473. Available: https://proceedings.neurips.cc/paper/2020/hash/53c04118df112c13a8c34b38343b9c10-Abstract.html
12. SVRTK: MIRTK based SVR reconstruction. Github; Available: https://github.com/SVRTK/SVRTK
13. Kainz B. fetalReconstruction: GPU accelerated source code for motion compensation of multi-stack MRI data. Github; Available: https://github.com/bkainz/fetalReconstruction
14. NiftyMIC: NiftyMIC is a research-focused toolkit for motion correction and volumetric image reconstruction of 2D ultra-fast MRI. Github; Available: https://github.com/gift-surg/NiftyMIC
15. Tustison NJ, Avants BB, Cook PA, Zheng Y, Egan A, Yushkevich PA, et al. N4ITK: improved N3 bias correction. IEEE Trans Med Imaging. 2010;29: 1310–1320.
16. Ranzini MBM, Fidon L, Ourselin S, Modat M, Vercauteren T. MONAIfbs: MONAI-based fetal brain MRI deep learning segmentation. arXiv [eess.IV]. 2021. Available: http://arxiv.org/abs/2103.13314
Figures