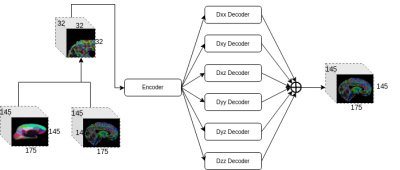0075
Reducing the impact of disrupted brain regions in Diffusion Tensor Imaging with inpainting1Brain and Mind Centre, University of Sydney, Camperdown, Australia, 2School of Computer Science, University of Sydney, Camperdown, Australia, 3Sydney Neuroimaging Analysis Centre, Camperdown, Australia
Synopsis
Keywords: Data Analysis, Diffusion Tensor Imaging
Diffusion Weighted Imaging (DWI) can be disrupted due to acquisition constraints or imaging artifacts, which can lead to unreliable scalar metrics calculated and valuable scans discarded as a result. To reduce the impact of disrupted brain regions in DTI, we adapted a deep learning DTI inpainting network to reconstruct the disrupted ROIs. We evaluated the performance of the method by calculating the Fractional Anisotropy errors according to each individual brain region. Experimental results show that the inpainting method can reconstruct the relevant clinical imaging information by mitigating the Fractional Anisotropy differences overall and in individual disrupted brain regions.Introduction
Diffusion Weighted Imaging (DWI) is a noninvasive medical imaging technique commonly used in neurological clinical research. A Diffusion Tensor Imaging (DTI) model can be fit from DWI to investigate and characterize microstructural information that can be summarised using different volumetric scalar metrics including Fractional Anisotropy (FA), Mean Diffusivity, and Axial Diffusivity1. Significant differences in scalar metrics can often be observed between healthy controls and patients in clinical studies and can be considered as biomarkers for particular neurological disorders. However, the DTI model can present errors due to acquisition constraints (lack of field of view) or external factors (motion artifacts, susceptibility distortions, and eddy currents). These errors can then lead to unreliable scalar metrics and valuable scans discarded as a result. To reduce the impact of disrupted brain regions in DTI, we adapted a deep-learning DTI inpainting network to reconstruct the disrupted regions of interest (ROIs). We evaluated the performance of the method by calculating the Fractional Anisotropy errors according to each individual region in the Desikan-Killiany Atlas2. Experimental results show that our method can reconstruct the disrupted ROIs with valid information in the DTIs and reduce the FA errors to a significantly lower level.Methods
100 different subjects were selected from the Human Connectome Project (HCP) database3 for the experiment. The HCP dataset consists of T1-weighted imaging and diffusion-weighted imaging acquired in a 3T Siemens “Connectom” Skyra scanner. The acquisition settings include high-resolution T1 acquisition (1.25 mm isotropic resolution, TR/TE = 5520/89.5 ms, and flip angle = 78°) and three diffusion weighted shells with b-values of 1000, 2000, and 3000 in the acquisition protocols of diffusion MRI, where each shell consists of 90 directions. Standard pre-processing steps were applied to the raw images to derive the corrected images in the same space4,5. DTIFIT from FSL package was then used to fit a DTI model from the corresponding processed DWIs. All the processed images had a size of 145 x 174 x 145, and 6 unique tensor values per voxel for the DTI volume. The Desikan-Killiany (DK) atlas was registered to the subject’s space.Tensor-wise Brain-aware Gate Network is a deep learning network specifically designed for inpainting disrupted regions (the details of the network are described elsewhere [6]). The network architecture is shown in Figure 1. Briefly, TW-BAG adopts 3D U-Net as its backbone and integrates two modules to learn a better representation of the disrupted ROIs. The vanilla convolutions are replaced by the brain-aware gate convolutions and 6 decoders are separated to learn each individual tensor coefficient from the encoder.
100 subjects were randomly split into 80 and 20 subjects for training and testing, respectively. Each unique tensor value was z-score normalized before being cropped into patches. The patch size was set to 32 x 32 x 32 as any region defined in DK atlas can be covered by this patch size. Each individual region was masked as 0 and its bounding box of 32 x 32 x 32 from the original DTI was fed into the network. The network was trained for 50 epochs using the L1 loss and Adam optimizer. The disrupted DTI values for a given ROI were then synthesized by setting the values of all the tensor coefficients in the DK atlas to 0. To evaluate the inpainting performance on a specific region, the fractional anisotropy was calculated in all the cortical surface regions from the DK atlas for the gold standard, the disrupted image, and the inpainted predictions for comparison.
Results
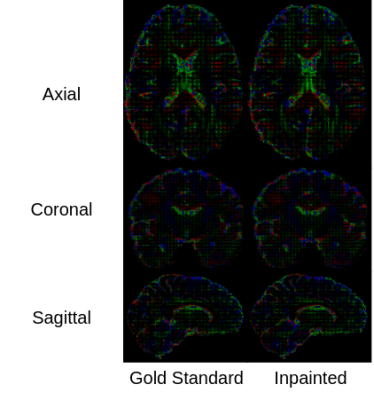
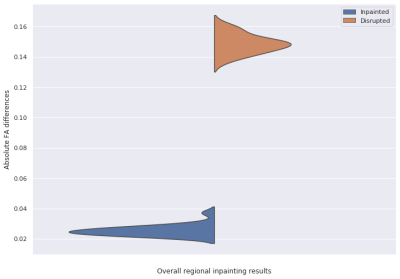
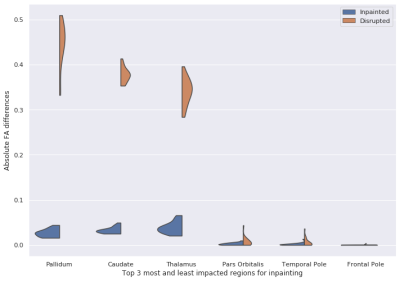
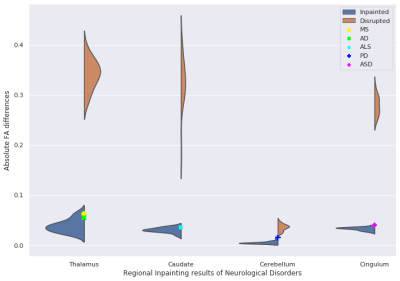
Figure 2 shows the visualization results of inpainted DTI and the gold standard in three different views. Figure 3 shows that the FA error of the disrupted DTIs is reduced from 0.1486 to 0.0237 after inpainting. The top 3 most and least impacted regions for inpainting are shown in Figure 4. To further investigate the clinical impact of TW-BAG, we collected several regions in the DK atlas due to their reported groupwise FA differences between controls and patients with the corresponding neurological disorders7-11. The mitigation of FA differences in the corresponding selected regions for the inpainted DTIs is shown in Figure 5. The FA gaps between controls and patients are highlighted with different markers.Discussion
Figure 3 and 4 demonstrate that TW-BAG is able to mitigate the FA differences bought by disrupted regions with a fair margin. Figure 5 shows that the measurement errors introduced by the disrupted regions could potentially impact the observed group differences between patients with corresponding neurological diseases and controls, where TW-BAG achieves satisfactory reconstruction performance from a clinical perspective by mitigating most of the FA errors to a lower level than the reported group differences. This can prevent erroneous conclusions in clinical studies and these studies could include subjects with disrupted DTIs by inpainting them instead of discarding the subject.Conclusion
We evaluated a deep learning-based inpainting approach specifically designed for reconstructing disrupted brain regions in DTIs. The experiment results suggest that the TW-BAG framework is able to reconstruct the missing clinical information in the disrupted ROIs from the valid information. Further clinical validation is required to explore the feasibility of using TW-BAG as a post-scanning tool for disrupted DTIs in clinical studies.Acknowledgements
The authors acknowledge the funding support by the Australia Medical Research Future Fund under Grant (MRFFAI000085).References
[1] Soares, J. M., Marques, P., Alves, V., et al., 2013. A hitchhiker's guide to diffusion tensor imaging. Frontiers in neuroscience, 7, 31.
[2] Desikan, R. S., Ségonne, F., Fischl, B., et al., 2006. An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. Neuroimage, 31(3), 968-980.
[3] Van Essen, D. C., Smith, S. M., Barch, et al., 2013. The WU-Minn human connectome project: an overview. Neuroimage, 80, 62-79.
[4] Jenkinson, M., Beckmann, C. F., Behrens, T. E., et. al., 2012. Fsl. Neuroimage, 62(2), 782-790.
[5] Fischl, B., 2012. FreeSurfer. Neuroimage, vol. 62, no. 2, pp. 774-781.
[6] Tang, Z., Wang, X., Zhu, L., et al., 2022. TW-BAG: Tensor-wise Brain-aware Gate Network for Inpainting Disrupted Diffusion Tensor Imaging. The International Conference on Digital Image Computing: Techniques and Applications (DICTA), 2022.
[7] Liu, Y., Duan, Y., Huang, J., et al., 2015. Multimodal quantitative MR imaging of the thalamus in multiple sclerosis and neuromyelitis optica. Radiology, 277(3), 784-792.
[8] Sharma, K. R., Sheriff, S., Maudsley, A., et al., 2013. Diffusion tensor imaging of basal ganglia and thalamus in amyotrophic lateral sclerosis. Journal of Neuroimaging, 23(3), 368-374.
[9] Ito, M., Watanabe, H., Kawai, Y., et al., 2007. Usefulness of combined fractional anisotropy and apparent diffusion coefficient values for detection of involvement in multiple system atrophy. Journal of Neurology, Neurosurgery & Psychiatry, 78(7), 722-728.
[10] Rose, S. E., Janke Phd, A. L., & Chalk, J. B., 2008. Gray and white matter changes in Alzheimer's disease: a diffusion tensor imaging study. Journal of Magnetic Resonance Imaging, 27(1), 20-26.
[11] Ikuta, T., Shafritz, K. M., Bregman, J., et al., 2014. Abnormal cingulum bundle development in autism: a probabilistic tractography study. Psychiatry Research: Neuroimaging, 221(1), 63-68.
Figures