0052
A novel ensemble deep learning neural network of multi-scale deep MRI features to localize the epileptogenic zone in pediatric epilepsy
Jeong-Won Jeong1,2, Min-Hee Lee1,2, Csaba Juhász1,2, and Eishi Asano1,3
1Pediatrics, Wayne State University, Detroit, MI, United States, 2the Translational Imaging Laboratory, Children's Hospital of Michigan, Detroit, MI, United States, 3Neurology, Children's Hospital of Michigan, Detroit, MI, United States
1Pediatrics, Wayne State University, Detroit, MI, United States, 2the Translational Imaging Laboratory, Children's Hospital of Michigan, Detroit, MI, United States, 3Neurology, Children's Hospital of Michigan, Detroit, MI, United States
Synopsis
Keywords: Neuro, Epilepsy, Ensemble deep learning, Localization of epileptogenic zone
We present a novel ensemble deep learning neural network of multi-scale deep MRI features that can non-invasively localize the epileptogenic zone (EZ) partially overlapping seizure onset zone (SOZ) and irritative zone (IZ) in children with medically intractable epilepsy. The presented network provided high balanced accuracy of 90% to predict SOZ and IZ in 3-fold cross-validation. It also yielded a new MRI marker of epileptogenicity providing a huge effect size between the ground-truth EZ and non-EZ (i.e., Cohen’s d = 2.73), suggesting its high translational value for accurately guiding invasive EEG practice to determine the boundaries of the EZ sitesIntroduction
Surgical resection of the epileptogenic zone (EZ)1 is an outstanding option for children whose seizures do not respond to medical therapy. Seizure onset zone (SOZ)2 and irritative zone (IZ, defined as the region generating interictal spikes)3 on invasive EEG (iEEG) are electrophysiological biomarkers used to prospectively localize the EZ in clinical practice. Only 50-70% of patients achieve seizure freedom following the resection based on semiology, scalp EEG, iEEG, MRI, and other available neuroimaging workups, often due to incorrect or incomplete localization of the EZ4. This study proposes a novel ensemble learning neural network5,6 combining a series of multi-scale residual networks (msResNet)7 to accurately localize the EZ sites by using their multi-scale deep features of clinical multi-modal MRI.Methods
We utilized the 3T multi-modal MRI clinically acquired from our retrospective archive that were originally utilized for our previous study7 (n=41 children with intractable epilepsy who underwent two-stage resective surgery and achieved long-term seizure freedom after surgery, age: 9.9±5.6 years old, 22 boys). A model cohort (n=24 children) was used using a 3-fold cross-validation to train and test the proposed ensemble learning neural network for accurate prediction of C1: SOZ, C2: IZ, and C3: Non-EZ in the end-to-end fashion (Fig. 1). A validation cohort (n=17 children) was then used to evaluate the reproducibility of the proposed network in an independent cohort. Briefly, the node-wise prediction was performed in the connectivity network, G = (Ωi=1-500, Ai=1-500,j=1-500), where Ωi defines the ith network node of the epileptogenic hemisphere in the Lausanne 2008 cortical parcellation atlas8 and Aij represents pair-wise white matter pathway edge connecting Ωi and Ωj (e.g., connectivity strength, the total count of connecting tracts normalized by the total volume of Ωi and Ωj). At each node Ωi, a surface laminar analysis7,9 was applied to extract a multi-modal MRI feature vector Xi sized by 1 x 1,800 consisting of I: relative intensity (RI) values of T1-weighted, T2-weighted, FLAIR, apparent diffusion coefficient (ADC), fractional anisotropy (FA) at two gray matter surfaces (outer/middle for cortical layer II and III), D: RI values at the deep white matter surface of FA, ADC, apparent fiber density (AFD), and C: DWI connectome (DWIC) profile (i.e., edge strengths Aij=1-500 sorted from the nearest to the farthest node). Three blocks of msResNet were separately pre-trained to extract three sets of deep feature vector ki that are most predictive of C1, C2, and C3. The resulting deep feature sets were combined in the framework of ensemble learning to predict three output probability values, [P(C1: SOZ), P(C2: IZ), P(C3: non-EZ)] from the given input vector Xi. Note that the proposed ensemble learning neural network consists of three msResNet blocks that are pre-trained to perform three binary classifications, 1) C1: SOZ vs. C3: non-EZ, 2) C1: SOZ vs. C2: IZ, 3) C2: IZ vs. C3: non-EZ, in the framework of ensemble learning that can predict three classes more accurately in the output layer. Finally, a new MRI marker of the epileptogenic likelihood εi was estimated by non-linearly combining two epileptogenic potentials, seizure onset likelihood μi (i.e., P(C1)) and spiking likelihood γi (i.e., P(C2)), after factorized by two exponents, a1 and a2 that weigh the individual contribution of SOZ and IZ to overall epileptogenicity according to their degrees of MRI abnormalities. An attention layer10 was used to identify specific MRI modalities that play the most predictive role for correct classification. Nodes surgically resected iEEG-defined SOZ (or IZ) and preserved iEEG-defined non-SOZ (or non-IZ), in which all patients achieved long-term seizure freedom, were treated as the ground-truth EZ and non-EZ, respectively.Results
The proposed ensemble network provided higher accuracy in correctly predicting three classes, C1-3 in both model and validation cohorts (90% and 80%, Fig 2), compared with the standalone msResNet (86% and 72%, Fig 2). It also yielded a huge effect size of εi (i.e., Cohen’s d = 2.73) to differentiate the ground-truth EZ from non-EZ (Fig. 3A) and identify that reduced FA of cortical layers (II and III) and increased DWIC profile are the most predictive markers to localize the iEEG-defined SOZ and IZ (Fig. 3B).Discussion
To the best of our knowledge, the present study is the first work reporting that a novel ensemble learning neural network of deep MRI features (intensity, diffusivity, connectivity) can predict the iEEG-defined SOZ and IZ more accurately and also identify the most predictive MRI markers that should be used to localize the EZ in current preoperative MRI protocol. This non-invasive prediction may help guide any type of invasive iEEG recording since it is solely based on the deep learning of preoperative MRI abnormalities, associated with iEEG abnormalities, that are often invisible on clinical MRIs. Furthermore, the proposed MRI-based epileptogenic likelihood can be used as a priori information to quantify the severity of seizure excitability.Conclusion
The findings of this study suggest a promise of clinical MRI-based ensemble deep learning approach that can make specific iEEG abnormalities (SOZ and IZ) more readily appreciable via intelligently recognizing and highlighting highly discriminative but subtle MRI abnormalities associated with specific iEEG abnormalities. Further investigation will provide important implications for applying the proposed deep learning-based preoperative evaluation to clinical presurgical workups.Acknowledgements
This research was supported by grants from the National Institutes of Health, R01 NS089659to J.J. and R01 NS064033 to E.A.References
- Rosenow F, Lüders H. Presurgical evaluation of epilepsy. Brain. 2001;124(9):1683-1700.
- Asano E, Juhász C, Shah A, et al. Role of subdural electrocorticography in prediction of long-term seizure outcome in epilepsy surgery. Brain. 2009;132(4):1038-1047.
- Hufnagel, A., Dümpelmann M, Zentner J, et al. Clinical relevance of quantified intracranial interictal spike activity in presurgical evaluation of epilepsy. Epilepsia. 2000;41(4):467-478.
- Hader WJ, Tellez-Zenteno, Metcalfe A, et al. Complications of epilepsy surgery: a systematic review of focal surgical resections and invasive EEG monitoring. Epilepsia. 2013;54(5):840-847.
- Budzik J. Many heads are better than one: The case for ensemble learning. KDnuggets September, 2019. https://www.kdnuggets.com/2019/09/ensemble-learning.html
- Jin LP, Dong J. Ensemble deep learning for biomedical time series classification. Comput Intell Neurosci. 2016;2016:6212684.
- Jeong JW, Lee MH, Kuroda N, et al. Multi-scale deep learning of clinically acquired multi-modal MRI improves the localization of seizure onset zone in children with drug-resistant epilepsy. IEEE J Biomed Health Inform. 2022 (Early access). https://doi.org/10.1109/JBHI.2022.3196330
- Daducci A, Gerhard S, Griffa A, et al. The connectome mapper: an open-source processing pipeline to map connectomes with MRI. PLoS One. 2012;7(12):e48121.
- Govindan RM, Asano E, Juhász C, et al. Surface‐based laminar analysis of diffusion abnormalities in cortical and white matter layers in neocortical epilepsy. Epilepsia. 2013;54(4):667-677.
- Xu H, Dong M, Lee MH, et al. Objective detection of eloquent axonal pathways to minimize postoperative deficits in pediatric epilepsy surgery using diffusion tractography and convolutional neural networks. IEEE Trans Med Imaging. 2019;38(8):1910-1922.
Figures
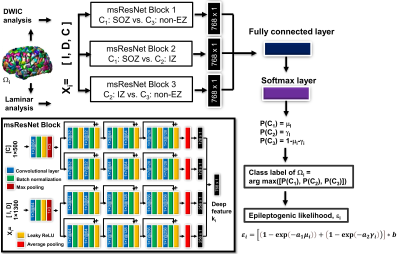
Figure
1. Detailed
architecture of the proposed ensemble deep learning neural network to deeply
mine the most discriminative deep features of three msResNet blocks, 1) C1:
SOZ vs. C3: non-EZ, 2) C1: SOZ vs. C2: IZ, 3) C2:
IZ vs. C3: non-EZ and predict three output class labels, C1:
SOZ, C2: IZ, and C3: non-EZ from a given input vector of
multi-modal MRI features, Xi = [I (i.e., RI values of T1-w, T2-w,
FLAIR, ADC, FA at two gray matter surfaces) D (i.e., RI values of FA, ADC, AFD
at a deep white matter surface), C (i.e., DIWC neighboring connectivity
profile)].
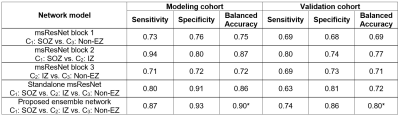
Figure 2. Balanced accuracy (i.e., arithmetic
mean of sensitivity and specificity) of the individual network model to predict
C1: SOZ, C2: IZ, and C3: non-EZ. An asterisk
indicates a maximum value.
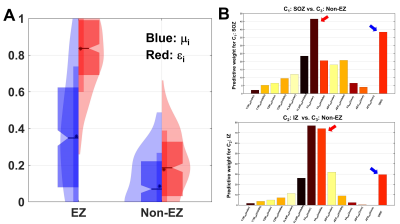
Figure 3. A. Violin and box plots (median/quartiles)
of μi (blue) and εi (red) evaluated from the ground-truth EZ and non-EZ. εi provided larger effect size than μi (i.e., Cohen’s d = 2.73 and 1.12 for ei and mi).
B. FA of the gray matter surface: FAGM (cortical layer II and
III, red arrow) and neighboring connectivity profile: DWIC (blue arrow) were
the most predictive to differentiate C1: SOZ and C2: IZ from
C3: non-EZ (msResNet Block 1 and 3, Fig. 1), suggesting the remarkable
presence of atypical changes in anisotropic integrity and neighboring connectivity
between the EZ and non-EZ.
DOI: https://doi.org/10.58530/2023/0052