0020
Improved 23Na Multi-Quantum Coherences MRI with simultaneous Cartesian Double-Half-Echo Sodium readouts1Computer Assisted Clinical Medicine, Heidelberg University, Mannheim, Germany, 2CRMBM, Aix-Marseille Université, Marseille, France
Synopsis
Keywords: Non-Proton, Multi-Contrast, Sodium
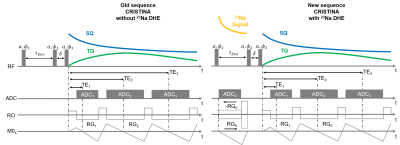
Sodium (23Na) MRI has received increased attention as a potential biomarker for disease states thanks to the advent of high and ultra-high field MRI. One asset from 23Na MRI is the distinction between single and triple quantum signal to provide new insights in tissue characterization. Hence, efficient sequences are needed that enable simultaneous 23Na and 23Na multi-quantum-coherences imaging to fully exploit the signal's potential. Therefore, we propose a new sequence utilizing a double-half-echo 23Na readout. Additionally, we propose to exploit this image further to drive the reconstruction of the low resolution MQC images.Cartesian TQ signal sampling requires three-pulse RF phase-cycling with a fixed time between the first two RF pulses to enable creation of the MQC. It has been shown that the evolution time can be used to insert additional readouts in between to obtain a conventional higher SNR 23Na image4. However, capturing this signal requires short echo times usually realized by utilization of half-radial sequences, for which the point-spread function differs from Cartesian echoes. Hence, we propose to leverage the Cartesian Double-Half-Echo (DHE) technique to capture a high quality sodium image in between the first two RF pulses.
In addition, the DHE sodium image can reach higher resolution and drive a super-resolution of the later MQC echoes. Shared information, such as edges, can be utilized by a prior total variation (TV) model to locally support the reconstruction of finer 23Na MQC images5.In this study, we leveraged a DHE Cartesian readout to obtain high-resolution sodium images that served to support super-resolution of the low SNR MQC images. The technique is demonstrated on phantom and in-vivo brain data acquired at 7T.
Methods
All measurements were performed on 7T MRI (Terra, Siemens Heathineers, Erlangen,Germany) with a bird-cage dual-tuned 23Na /1H head coil (QED, Mayfield Village OH, USA). 23Na MQC images were obtained by utilizing the CRISTINA sequence6 and an adapted version that features an additional Cartesian Double-Half Echo7 23Na readout between the first two RF pulses, see Fig.1. The following parameters were used:In phantom: 200x200x200mm3, matrix size 40x40x10, $$$\tau$$$ =10ms, BW=330Hz/px,TE/ $$$\Delta$$$TE/NTE= 1.11/4.2ms/10, TR=150ms, 2 averages resulting inTA=2x20 minutes. Sodium readout matrix size 60x60x10, TE=0.7ms, 7 averages.
For brain in-vivo, two volunteers (28+/-1, f, m): FoV 230x230x160mm3, matrix size 28x28x12, $$$\tau$$$=10ms, BW=330Hz/px, TE/$$$\Delta$$$TE/NTE= 1.11/4.2ms/10, TR=160ms resulting in TA=2x25min. Sodium readout matrix size 60x60x10, TE=0.7ms, 7 averages. 2D FLASH matrix size 210x210x32, TA ~1min.
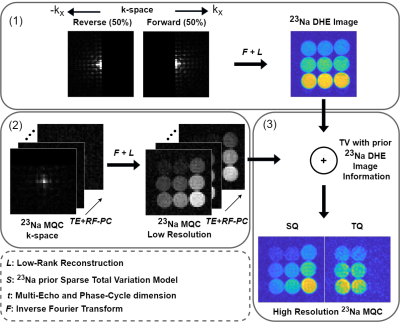
Image reconstruction: First, the sodium raw data from the readout between 1st and 2nd RF pulse was processed by utilizing low-rank matrix completion that combines both halves of k-space7,8. Afterwards, 23Na MQC raw data were processed by first computing a low-rank approximation that exploits coherences in the data along the temporal and phase-cycle dimensions. Then, 3D Total Variation (TV) was used to share information, such as tissue boundaries, between 23Na DHE and MQC readouts (Fig.2).
Results
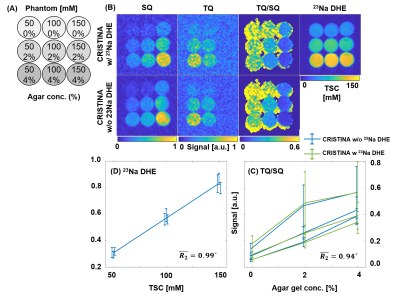
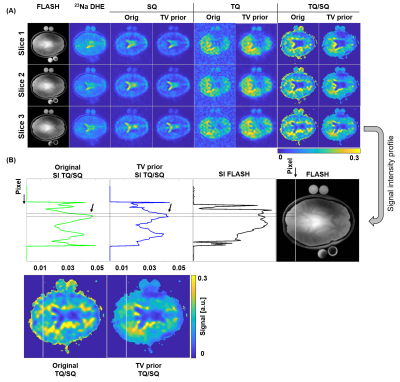
The phantom study showed that the new sequence provides reliable SQ and TQ images and despite, also a Cartesian UTE sodium image. Linear regression revealed accurate signal intensity versus known NaCl concentration for the DHE image (Fig.3).In-vivo brain 23Na images showed the typical sodium contrast in the brain, with additionally lower resolution SQ and TQ images acquired simultaneously (Fig.4). The 23Na DHE image contrast is in line with the contrast provided by the SQ image exhibiting highest signal in the CSF compartment. However, the TQ signal was lowest in CSF, yielding accurate TQ/SQ ratios with clear tissue delineation.
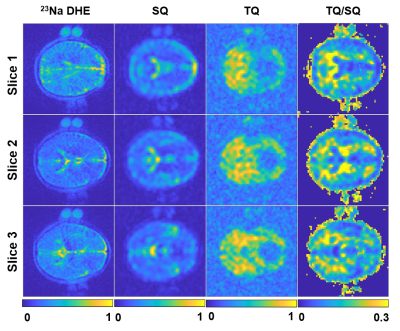
For in-vivo brain data, enhanced resolution for MQC images was achievable by enhancing tissue boundaries (Fig.5). The higher SNR sodium image provides thus useful information to also improve 23Na MQC image quality, without the need of acquiring an additional scan. Furthermore, the 23Na DHE and the 23Na MQC images are highly correlated in regards to sampling (Cartesian) but also image contrast, since tissue boundaries are sodium-density weighted and not 1H.
Discussion and Conclusion
In-depth study is warranted that shows the potential benefits of quantifying TSC on the DHE images. It is expected to reduce the influence of T2* bias, which is present in the SQ image, and could thus be useful for increased accuracy in TSC quantification. Additionally, future work might include optimizing the DHE readout to utilize multiple gradient echoes for T2* short component quantification, and jointly estimate the T2* long component from MQC images.Conclusively, we have demonstrated that the double-half-echo technique enables to capture Cartesian UTE 23Na images inbetween the preparation time of MQC. Furthermore, it has been shown that the high SNR sodium image information can be utilized to improve the reconstruction of low resolution 23Na MQC images. Hence, the new sequence with the new reconstruction pipeline offers great potential to be used in a clinical setting to fully exploit the potential of 23Na MQC MRI.
Acknowledgements
The author received financial support for the research by ISMRM research exchange grant program and PROCOPE Mobility 2022.References
1. Huhn K, Engelhorn T, Linker RA, Nagel AM. Potential of Sodium MRI as a Biomarker for Neurodegeneration and Neuroinflammation in Multiple Sclerosis. Front Neurol. 2019 Feb.
2. Hoesl, M.A., Kleimaier, D., Hu, R., Malzacher, M., Nies, C., Gottwald, E. and Schad, L.R. (2019), 23Na Triple-quantum signal of in vitro human liver cells, liposomes, and nanoparticles: Cell viability assessment vs. separation of intra- and extracellular signal. J. Magn. Reson. Imaging, 50: 435-444.
3. LaVerde, G., Nemoto, E., Jungreis, C., Tanase, C. and Boada, F. (2007), Serial triple quantum sodium MRI during non-human primate focal brain ischemia. Magn. Reson. Med., 57: 201-205.
4. Fiege DP, Romanzetti S, Mirkes CC, Brenner D, Shah NJ. Simultaneous single-quantum and triple-quantum-filtered MRI of 23Na (SISTINA). Magn Reson Med. 2013 Jun;69(6):1691-6.
5. Licht, C., Rapacchi, S., Schad, L.R., An iterative algorithm to resolve high-resolution 23Na Multi-Quantum Coherences MRI from prior 1H constraints. ISMRM 2022 Joint Annual Meeting, Digital Poster, 2022.
6. Hoesl, M.A.U., Schad, L.R., Rapacchi, S.,Volumetric 23Na Single and Triple-Quantum Imaging at 7T: 3D-CRISTINA, Zeitschrift für Medizinische Physik, Volume 32, Issue 2, 2022, Pages 199-208.
7. Bydder M, Ali F, Ghodrati V, Hu P, Yao J, Ellingson BM. Minimizing echo and repetition times in magnetic resonance imaging using a double half-echo k-space acquisition and low-rank reconstruction. NMR Biomed. 2021 Apr;34(4):e4458. doi: 10.1002/nbm.4458. Epub 2020 Dec 9. PMID: 33300182; PMCID: PMC7935763.
8. Shin PJ, Larson PE, Ohliger MA, Elad M, Pauly JM, Vigneron DB, Lustig M. Calibrationless parallel imaging reconstruction based on structured low-rank matrix completion. Magn Reson Med. 2014 Oct;72(4):959-70.
Figures