0013
Expanding interleaved MRI-DMI to 3D SWI (Susceptibility Weighted Imaging) : A truncated acquisition approach1Yale University, New Haven, CT, United States
Synopsis
Keywords: Deuterium, Deuterium, DMI
Interleaved MRI-DMI (Deuterium Metabolic Imaging) is a transformative approach to obtain multi-contrast anatomic MRI and metabolic information in parallel with time efficiency. In this work, we expand the toolkit of interleaving 2H DMI with 1H MRI, by using a newly developed interleaved 3D SWI-DMI sequence that relies on acquiring a truncated 2H signal. Acknowledging the limitations of fitting truncated 2H signals independently, we propose a method to combine truncated FID with fully-sampled acquisitions to improve DMI sensitivity. The updated interleaved MRI-DMI protocol with 3D SWI-DMI was demonstrated on a patient with a brain tumor.Introduction
Clinical assessment of neurological diseases can benefit from metabolic information in addition to the standard anatomical MRI routine. A number of MRS(I) techniques, based on 1H, 2H, 13C, HP-13C or 31P can be used to characterize metabolism, among which Deuterium Metabolic Imaging (DMI)1 is the most recent, and a promising method for clinical translation due to its robustness and simplicity. Despite the advantages, a sequential acquisition of MRI and DMI will significantly prolong an MRI scan session. To overcome this restriction, we developed a method to acquire DMI and MRI in parallel by interleaving the 2H sequence elements into the 1H MRI delays with a demonstration provided for FLAIR-DMI.2 Since then, we have expanded the principle to a variety of MRI scans (MP-RAGE, 2D SWI, T2W), covering a complete clinical protocol.3 However, not all MRI methods have sufficiently long delays to accommodate the insertion of DMI. 3D-SWI4 is one example where the short TR (30-50ms) and long TE (15-25ms) does not allow the insertion of a full-length DMI acquisition (80ms). Here we explore the use of truncated DMI acquisitions in interleaved MRI-DMI using simulated data, and in vivo on a patient with a brain tumor.Methods
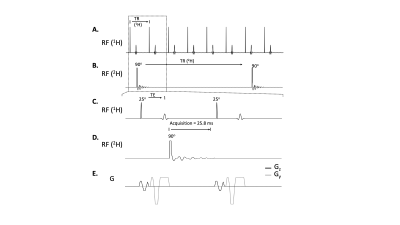
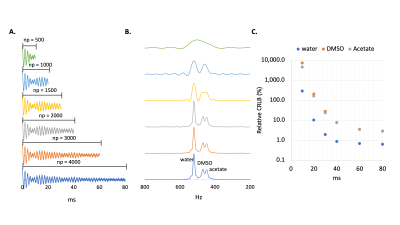
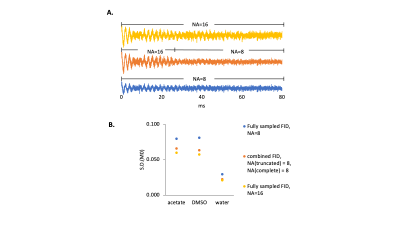
Interleaved 3D SWI-DMI sequence. The interleaved 3D SWI-DMI RF pulse sequence scheme is shown in Figure 1. SWI (TR/TE=52.4/25ms, 256x192x64 matrix 256x192x128mm3) was interleaved with DMI (13x9x11 matrix over 260×180×220mm3, spherical k-space encoding). Approximately 25.8ms of 2H FID acquisition window can be achieved (np=1290, sw=50kHz) without further prolonging the 1H TR. A 2H TR of 314ms, consistent with previously developed interleaved MRI-DMI sequences5, was maintained by interleaving one 2H excitation-acquisition for every six 1H excitations. For a total SWI acquisition time of 10min and 43s , 4 repeats of truncated DMI were acquired.2H FID simulation. To evaluate the effect of FID signal truncation on quantifying the 2H signal, an FID signal containing water, DMSO and acetate resonances was simulated as damped sinusoidal functions with linewidths of 10, 15 and 15Hz, respectively and various levels of Gaussian distributed noise. FIDs were calculated from a standard acquisition length of 80ms, displaying minimal truncation effects, down to a heavily truncated period of only 10ms. The simulated FIDs were fitted via Linear Combination Modeling (LCM) as previously described2, assuming a fixed chemical shift for each compound. Uncertainty in the fitting parameters was evaluated with Cramer-Rao Lower Bounds6 (CRLB). To obtain a combined FID, truncated signal (30ms, 8 averages) was zero-filled and summed directly with the fully-sampled signal (50ms, 8 averages). Following average-weighted FID fitting, the fitting accuracy was determined using Monte-Carlo simulation and represented using standard deviation (S.D.) of fitted M0.
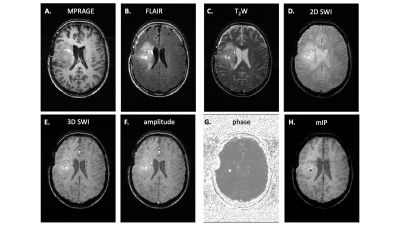
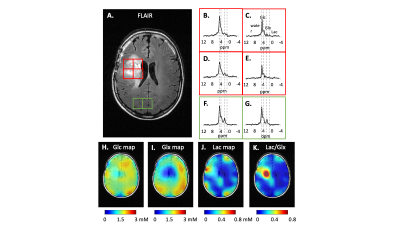
Patient study. The previously developed interleaved MRI-DMI routine, including FLAIR-, MP-RAGE-, T2W-, and 2D SWI-DMI, was combined with the 3D SWI-DMI sequence to acquire multi-contrast MRI and DMI on a brain tumor patient, 75 min after oral intake of [6,6’-2H2]-glucose as described previously1. For a total acquisition time of 40 minutes, 9 repeats of complete DMI and 4 repeats of truncated DMI were acquired in parallel to the MRIs. The complete and truncated DMI were combined to generate metabolic maps following spectral fitting.
Results
To understand the truncation effect on 2H signal, FIDs were simulated with various acquisition length (Figure2A). Following Fourier transform, severe truncation artefacts and decreased spectral resolution were observed on the spectra as data acquisition shortened (Figure2B). A 10-30 fold increase of relative CRLB was observed on the peaks as the signal acquisition shortens from 80ms to 30ms (Figure2C). The interleaved 3D SWI-DMI allows an acquisition of 2H signal for 25.8ms (1290 data points over 50KHz) (Figure1). As suggested by the simulation (Figure2), the truncated data cannot be reliably fitted. However, truncated 2H FIDs can be combined with complete FIDs acquired during other interleaved MRI-DMI sequences to improve overall 2H signal sensitivity (Figure3A). The combined FID has non-Gaussian distributed noise and the errors cannot be quantified using CRLB. We therefore performed Monte-Carlo simulations to evaluate the error of fitting. The standard deviation of fitted M0 indicated a 20%-32% increase of fitting accuracy when 8 averages of truncated FID were combined with equal averages of fully-sampled FID. Although this improvement is less than combining the same number of fully-sampled FIDs (40% increase), the addition of truncated FIDs to the total number of averages still contributes significantly to increasing the overall SNR of the complete DMI dataset (Figure 5B). 3D SWI-DMI was combined with the previously developed interleaved MRI-DMI protocol3 and applied on a brain tumor patient. The 40-minute MRI-DMI protocol successfully acquired MRIs (Figure4) with expected contrast in addition to 9 averages of complete DMI and 4 averages of truncated DMI (Figure5). DMI combining fully-sampled and truncated signal (Figure5) was free from artefacts and can be used to reliably detect aberrant glucose metabolism in brain tumors.Discussion
We designed and implemented interleaved 3D SWI-DMI that acquires 3D SWI and truncated DMI in parallel. An approach to incorporate truncated FID with fully-sampled 2H signal was evaluated and its benefits on improving DMI sensitivity was demonstrated. The inclusion of truncated DMI acquisition further expands the flexibility of the interleaved 1H/2H methodology to accommodate an even wider range of MRI sequences.Acknowledgements
This research was funded, in part, by NIH grant NIBIB R01-EB025840.References
1. De Feyter HM, Behar KL, Corbin ZA, et al. Deuterium metabolic imaging (DMI) for MRI-based 3D mapping of metabolism in vivo. Science Advances. 2018;4(8):eaat7314.
2. Liu Y, De Feyter HM, Fulbright RK, McIntyre S, Nixon TW, de Graaf RA. Interleaved fluid-attenuated inversion recovery (FLAIR) MRI and deuterium metabolic imaging (DMI) on human brain in vivo. Magn Reson Med. 2022.
3. Liu Y, De Feyter HM, Fulbright RK, McIntyre S, Nixon TW, de Graaf RA. A Multi-Sequence, Interleaved MRI-DMI Protocol for Human Brain. 2022.
4. Haacke EM, Xu Y, Cheng YCN, Reichenbach JR. Susceptibility weighted imaging (SWI). Magnetic Resonance in Medicine: An Official Journal of the International Society for Magnetic Resonance in Medicine. 2004;52(3):612-618.
5. Liu Y, De Feyter HM, McIntyre S, Nixon TW, de Graaf RA. Interleaved MRI and DMI on human brain in vivo. ISMRM; 2021.6. Cavassila S, Deval S, Huegen C, van Ormondt D, Graveron-Demilly D. Cramér-Rao bounds: an evaluation tool for quantitation. NMR Biomed. 2001;14(4):278-283.
Figures