0003
Automated MR Image Prescription of the Liver using Deep Learning: Development, Evaluation & Prospective Implementation1Departments of Medical Physics, Radiology, University of Wisconsin, Madison, Madison, WI, United States, 2Department of Electrical and Computer Engineering, University of Wisconsin, Madison, Madison, WI, United States, 3Department of Radiology, University of Wisconsin, Madison, Madison, WI, United States, 4Department of Radiology and Nuclear Medicine, Universität zu Lübeck, Lübeck, Germany, 5Departments of Medical Physics, Radiology, Medicine, Emergency Medicine, Biomedical Engineering, University of Wisconsin, Madison, Madison, WI, United States, 6Departments of Medical Physics, Radiology, Electrical and Computer Engineering, Biomedical Engineering, University of Wisconsin, Madison, Madison, WI, United States
Synopsis
Keywords: YIA, Liver
This work developed a novel automated AI-based method for liver image prescription from a localizer and evaluated it in a large retrospective patient cohort (1,039 patients for training/testing), across pathologies, field strengths, and against radiologists’ inter-reader reproducibility performance. AI-based 3D axial prescription achieved a S/I shift of <2.3 cm compared to manual prescription for 99.5% of test dataset. The AI method performed well across all sub-cohorts and better in 3D axial prescription than radiologists’ inter-reader reproducibility performance. We successfully implemented the AI method on a clinical MR system, which demonstrated robust performance across localizer sequences.Introduction
To enable high-value MRI of the liver with improved workflow, efficiency, and reproducibility, automated image prescription is needed1,2. Previous efforts3,4 relied on traditional image processing tools, with limited performance and validation. However, reliable image prescription for the liver remains an unmet need. Artificial intelligence (AI) methods, as reported in other organs5-7, may address this need for liver MRI. Therefore, the purpose of this study was to develop AI-based fully automated prescription for liver MRI8.Methods
Data: In this IRB-approved study, data (localizer images) from 1,039 patient exams (Figures 1-2) were retrieved retrospectively, with a waiver of informed consent.Manual labeling and inter-reader reproducibility: Six board-certified abdominal radiologists manually annotated seven classes of labels using bounding boxes that span the liver, torso, and arms to enable liver image prescription in any orientation (Figure 1). Inter-reader reproducibility was evaluated.
Training: We trained a convolutional neural network (CNN) based on the YOLOv3 architecture for detection and classification of the aforementioned bounding boxes. Different training dataset sizes (5%-100% of the 831 training datasets) were evaluated. Finally, a shallower network (YOLOv3-tiny) was also trained to determine the tradeoff between inference speed and performance.
3D image prescription: Based on the detected bounding boxes for each slice in the localizer, image prescription for whole-liver acquisition in each orientation was calculated as the minimum 3D bounding box needed to cover all the labeled 2D bounding boxes in the required volume (Figure 1). For example, 3D axial prescription covers the torso in A/P, R/L dimensions and the liver in S/I.
Evaluation: The performance of automated 2D object detection for each of the seven classes of labels was measured by intersection over union (IoU). To evaluate the performance of subsequent automated 3D prescription, mismatch with manual prescription for each of the six edges of the 3D box was calculated. Performance was evaluated on 20% of datasets across patients, pathologies, field strengths and sequences, with increasing training sizes, and against inter-reader reproducibility results.
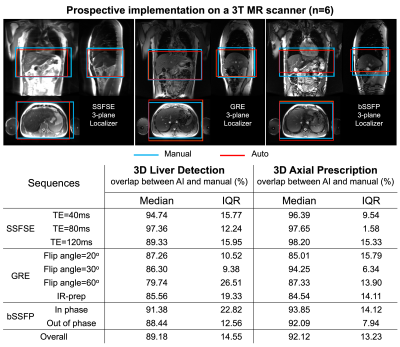
Online implementation & prospective study: The automated prescription was implemented on a 3T MR system (GE MR750), and evaluated prospectively on six healthy volunteers using multiple localizer pulse sequences (SSFSE, fGRE, bSSFP) and acquisition parameters.
Results
The resulting code and CNN weights are available at https://doi.org/10.5281/zenodo.7391054.AI-based liver prescription for 3D liver detection and 3D axial prescription maintained high performance across sub-cohorts in age, sex, BMI, pathology, and acquisition field strength and sequence (Figure 2). High overlap (>91%) between AI and manual labeling for 3D liver detection and 3D axial prescription was observed across all sub-cohorts.
Figure 3 shows the accuracy of 2D (a) and 3D detection (b), and prescription (c-e). The histograms of IoU for each of the seven classes are qualitatively similar, with IoU median >0.91 and interquartile range <0.09. For 3D liver detection, 93% ± 9% of the manual volume was included in the automated prescription, while 3D axial prescription had 97% ± 3% inclusion, 3D coronal and sagittal prescription had 96% ± 6% and 95% ± 7% inclusion respectively. The shift in 3D axial prescription was less than 2.3 cm in the S/I dimension for 99.5% of the test datasets.
The performance of AI-based prescription improved with training dataset size (Figure 4), and achieved performance comparable or superior to inter-reader reproducibility for 3D axial prescription. For axial prescription, the inter-reader reproducibility study yields a buffer of 2.4cm in the S/I dimension which would cover 99.5% of the test patients, on par with the AI-based method.
We successfully implemented the AI-based automated image prescription method with the full and tiny YOLOv3 networks on an MRI system at our site. Automated image prescription for one three-plane localizer on a CPU in the console required ~10 seconds with the full network and ~3 seconds with the tiny network. Both networks showed promising prospective performance (similar to that of the retrospective study) with no significant difference (p = 0.06) across the three localizer sequences acquired (Figure 5) except fGRE with IR-prep. AI-based prescription performed best when using spin-echo localizers.
Discussion
In contrast to previous methods based on traditional object detection3, the proposed AI-based prescription method uses conventionally acquired localizers with no additional scan needed, allows for prescription in any orthogonal direction, and has been tested in 200+ patients with a higher detection accuracy. Additional coverage considerations in each orientation can easily be included subsequently. For example, the small S/I shift in axial prescription indicates that the addition of narrow safety margins would ensure complete liver coverage in effectively all patients. This study has several limitations. Due to the limited number of failure cases in the testing dataset, the specific causes of occasional detection failures remains unclear. This study relied on data from a single center and a single vendor; the prospective pilot study on a clinical MR system had a small number of healthy subjects. Studies involving multi-center, multi-vendor datasets for retrospective and prospective validation are ongoing.Conclusion
This work demonstrated excellent performance of AI-based automated liver image prescription across patients, pathologies, and clinically relevant acquisition settings, as well as successful scanner implementation. Overall, the proposed AI method has the potential to standardize liver MR prescription.Acknowledgements
The authors would like to thank Daryn Belden, Wendy Delaney, and Prof. John Garrett from UW Radiology for their assistance with data retrieval, and Dan Rettmann, Lloyd Estkowski, Naeim Bahrami, Ersin Bayram, and Ty Cashen from GE Healthcare for their assistance with implementation of our AI-based liver image prescription on one of the GE scanners at the University of Wisconsin Hospital. The authors acknowledge support from the NIH (R01-EB031886). The authors also wish to acknowledge GE Healthcare and Bracco who provide research support to the University of Wisconsin. Dr. Oechtering receives funding from the German Research Foundation (OE 746/1-1). Dr. Reeder is a Romnes Faculty Fellow and has received an award provided by the University of Wisconsin-Madison Office of the Vice Chancellor for Research and Graduate Education with funding from the Wisconsin Alumni Research Foundation.
References
1. Itti L, Chang L, Ernst T. Automatic scan prescription for brain MRI. Magn Reson Med. 2001;45(3):486-494. doi:https://doi.org/10.1002/1522-2594(200103)45:3<486::AID-MRM1064>3.0.CO;2-#
2. Lecouvet FE, Claus J, Schmitz P, Denolin V, Bos C, Berg BCV. Clinical evaluation of automated scan prescription of knee MR images. J Magn Reson Imaging. 2009;29(1):141-145. doi:https://doi.org/10.1002/jmri.21633
3. Goto T, Kabasawa H. Automated Scan Prescription for MR Imaging of Deformed and Normal Livers. Magn Reson Med Sci. 2013;12(1):11-20. doi:10.2463/mrms.2012-0006
4. Goto T, Kabasawa H. Automated Positioning of Scan Planes and Navigator Tracker Locations in MRI Liver Scanning. Med Imaging Technol. 2010;28(4):245-251. doi:10.11409/mit.28.245
5. GE Healthcare’s AIRxTM Tool Accelerates Magnetic Resonance Imaging. Intel. Accessed December 15, 2020. https://www.intel.com/content/www/us/en/artificial-intelligence/solutions/gehc-airx.html
6. Barral JK, Overall WR, Nystrom MM, et al. A novel platform for comprehensive CMR examination in a clinically feasible scan time. J Cardiovasc Magn Reson. 2014;16(Suppl 1):W10. doi:10.1186/1532-429X-16-S1-W10
7. De Goyeneche A, Peterson E, He JJ, Addy NO, Santos J. One-Click Spine MRI. 3rd Med Imaging Meets NeurIPS Workshop Conf Neural Inf Process Syst NeurIPS 2019 Vanc Can.
8. Geng R, Buelo CJ, Sundaresan M, Starekova J, et al. Automated MR image prescription of the liver using deep learning: Development, evaluation, and prospective implementation. Journal of Magnetic Resonance Imaging. 2022 Dec 30.
9. Redmon J, Divvala S, Girshick R, Farhadi A. You Only Look Once: Unified, Real-Time Object Detection. ArXiv150602640 Cs. Published online May 9, 2016. Accessed December 15, 2020. http://arxiv.org/abs/1506.02640
10. Pang S, Ding T, Qiao S, et al. A novel YOLOv3-arch model for identifying cholelithiasis and classifying gallstones on CT images. PloS One. 2019;14(6):e0217647. doi:10.1371/journal.pone.0217647
Figures
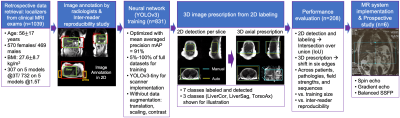
Summary of data retrieval, annotation, training, prescription, evaluation, and scanner implementation. Manual labeling involved 7 localization regions and was evaluated by inter-reader reproducibility. A CNN for object detection was trained with 80% datasets. Minimum 3D box needed to cover labeled 2D boxes in each view was used to obtain 3D prescription. Evaluation of 2D and 3D boxes was done in 20% datasets across patients and pathologies. We successfully implemented the method on a clinical MR system and conducted a prospective study with 6 volunteers across sequences.
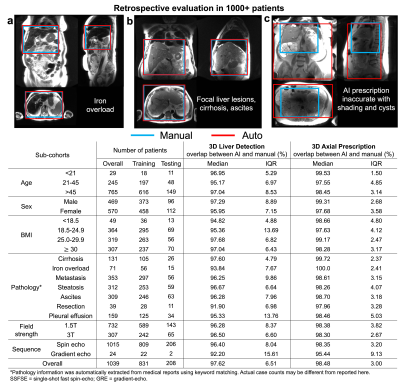
In most cases, the liver volume was covered accurately by automated prescription, including in patients with iron overload (a), focal lesions, cirrhosis and ascites (b). Inaccurate automated object detection was observed for patients with multiple renal cysts (c) due to dielectric shading. Distribution of patient datasets in age, sex, BMI, pathologies, acquisition field strength and sequence is shown in the table. Overlap between AI and manual labeling for 3D liver detection and axial prescription was high (>91%) across all categories.
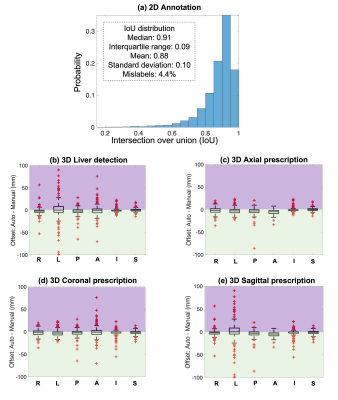
Accuracy of 2D annotation (a), 3D liver detection (b), and image prescription (c-e). IoU histograms for all classes are qualitatively similar, with IoU median >0.91 and interquartile range <0.09. In (b-e), x axis shows the 6 edges: right (R), left (L), posterior (P), anterior (A), inferior (I), superior (S); y axis shows difference between automated and manual volumes (0: perfect alignment; green areas: AI covering more volume; purple: missed volume). All boxes are tight around 0. The shift in 3D axial prescription was less than 2.3 cm in S/I dimension for 99.5% of test datasets.
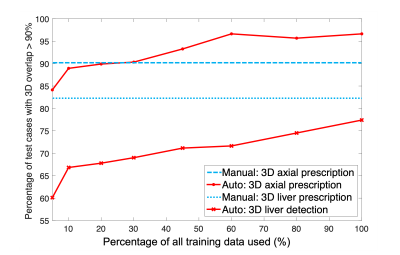
As training size increased, the percentage of test cases with high overlap (>90%) in 3D between AI and manual prescription increased for 3D liver detection and axial prescription. AI performance for 3D axial prescription plateaued after training with 500 patients' datasets (60% of training data). AI performance for 3D liver detection approached but never reached radiologists' inter-reader reproducibility performance. Training with at least 250 datasets (30% of training data), AI-based 3D axial prescription performed better than (manual) inter-reader reproducibility.