Osprey - A guide for a modern MRS analysis for everyone
Helge Jörn Zöllner1,2
1Russell H. Morgan Department of Radiology and Radiological Science, The Johns Hopkins University School of Medicine, Baltimore, MD, United States, 2Kennedy Krieger Institute, F.M. Kirby Research Center for Functional Brain Imaging, Baltimore, MD, United States
1Russell H. Morgan Department of Radiology and Radiological Science, The Johns Hopkins University School of Medicine, Baltimore, MD, United States, 2Kennedy Krieger Institute, F.M. Kirby Research Center for Functional Brain Imaging, Baltimore, MD, United States
Synopsis
Osprey is an open-source MRS analysis package streamlining uniform pre-processing, linear-combination modeling, tissue correction and quantification adhering to recent consensus guidelines. It provides an end-to-end analysis solution for the broader MRS community including application-focused researchers, and quick development benchmarking for methodologists. The software tutorial will cover a general overview of the software’s capabilities and demonstrate easy translation to the user’s own research questions. Here, we present Osprey’s modular capabilities for quick method development – showing published example studies – and translation into application research and teaching using Osprey’s job file system and example data.
Introduction
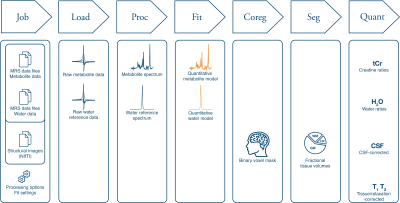
Recent expert consensus emphasizes the need for methodological harmonization in 1H-MRS to achieve reproducible and transparent research. Three key analysis components in the workflow of 1H-MRS following acquisition are preprocessing, spectral analysis and quantification. However, those steps are not widely standardized and typically require a combination of (potentially error prone) legacy code and third-party modeling software. Osprey seamless integrates preprocessing, linear-combination modeling (LCM), quantification, and data visualization in a single open-source workflow following recent consensus (Figure 1 & Figure 2). Because of its modular structure, Osprey allow for quick benchmarking of new methods into the pipeline, and can be immediately used for application-oriented research and teaching purposes.Description
Osprey is structured into seven modules which are executed according to a job file definition provided by the user. This file specifies input data (compatible with the BIDS organizational structure) and various processing and modeling options. The analysis pipeline includes loading of vendor-native raw data (also supporting DICOM and NIfTI-MRS), preprocessing, linear-combination modeling, coregistration of the MRS voxel and tissue segmentation, and finally quantification. It also allows to directly investigate spectra and quantitative outcome with various overview visualizations including cohort-mean spectra and raincloud plots of estimated parameters.The tutorial will demonstrate the different analysis stages, provide examples on how to set up job files, and explain the information provided by the user-friendly GUI. Further, the modular capabilities and its potential applications to quickly benchmark new methods will be showcased.