From Image Contrast to Quantitative Imaging
Tobias C Wood1
1King's College London, London, United Kingdom
1King's College London, London, United Kingdom
Synopsis
This talk will provide a grounding in how to turn standard MR contrast images into quantitative maps. Such maps represent meaningful physical and biological parameters with known units. This provides new information on a voxel-wise level with which to base clinical diagnoses. This development can be seen as part of the continuing development of MRI as a field, turning the scanner from a fancy camera into a precise scientific measuring instrument.
Target Audience
MR Physicists, researchers and clinicians interested in improving analysis and diagnosis by basing it on quantitative tissue properties instead of simple contrasts.Purpose
What is going on inside a voxel? Why can the same tissue appear hypo-intense in one sequence and hyper-intense in another? Can we use an MR scanner to actually measure tissue, instead of only taking a picture of it?A standard MR image is only qualitative, meaning that we cannot assign any significance to the intensity value in a particular voxel. We can only make relative comparisons between voxels - i.e. that one is brighter or darker than another. This means that diagnoses must be made by comparing image regions to each other, often across several different contrasts that highlight different aspects of pathology.
Quantitative MR imaging (qMRI) gives meaning to individual voxel values by making them represent physical quantities such as relaxation times or diffusion coefficients, with known units [1, 2]. This make it possible to test whether individual voxels are abnormal - a significant step forward for helping clinicians make informed diagnoses.
This talk will provide an introduction to the basic methodology of common qMRI techniques, discuss common pitfalls, and present potential analysis methods. This foundation will be set within a review of the evolution of MRI, from the early days where simply acquiring an image with sufficient contrast was a major technical achievement, to cutting edge methods where the dynamics of the MR signal can be exploited to yield rich quantitative information in the time of a normal scan.
Talk Outline
How to Turn Contrasts Into Parameters- The signal in any voxel is determined by the choice of sequence and a combination of tissue and sequence parameters. We can manipulate the contrast of images by manipulating the sequence parameters.
- If we have an adequate model of the MR signal, then by acquiring multiple images with different sequence parameters we can extract the tissue parameters, making quantitative maps.
- Linking biological parameters with physically measurable quantities is difficult and an active area of research.
- In addition to the parameters we are interested in, there are often additional parameters that must be modelled to adequately explain the signal. These include nuisance magnetic fields such as B0 and B1 inhomogeneity.
- Quantitative images need scans with known, fixed sequence parameters. Commercial scanners, through pre-scans or adjustments, actively fight against this. Knowing how to disable these is absolutely essential in quantitative imaging.
- qMRI allows interesting new analysis methods such as personalised pathology maps - comparing each voxel in the brain to a known distribution of values specific to that location in the brain [3, 4].
- However a significant unsolved problem in qMRI is adequate registration and alignment methods that do not sacrifice anatomical precision from high-resolution datasets.
- Classic qMRI methods rely on the acquisition of multiple images. Modern methods such as finger-printing treat each TR as individual data points, and through advanced reconstruction methods reconstruct multiple quantitative images from scans that are comparable to traditional qualitative images [5].
- This development can be seen as part of a historical trend in MRI from simple low-resolution images to high-resolution, highly informative, precise quantitative measurements.
Acknowledgements
Gareth Barker and Emil Ljungberg for many useful conversations.References
- Quantitative MRI of the Brain, 2nd Edition. CRC Press 2018
- Quantitative Magnetic Resonance Imaging, 1st Edition. Elsevier 2020
- Bonnier, G. et al. Personalized pathology maps to quantify diffuse and focal brain damage. NeuroImage: Clinical 21, 101607 (2019)
- Piredda, G. F. et al. Quantitative brain relaxation atlases for personalized detection and characterization of brain pathology. Magnetic Resonance in Medicine (2019)
- Wang, X., Tan, Z., Scholand, N., Roeloffs, V. & Uecker, M. Physics-based reconstruction methods for magnetic resonance imaging. Phil. Trans. R. Soc. A. 379, 20200196 (2021).
Figures
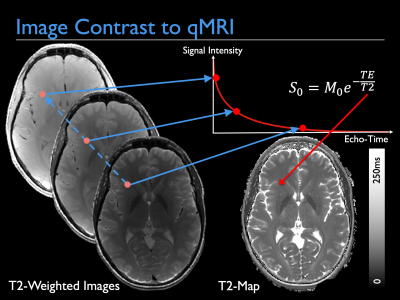
qMRI, here illustrated with T2 mapping, is classically achieved by acquiring images with different sequence parameters and then fitting a signal model to each voxel in order to extract the tissue parameters. This talk will describe how to do this, and recent advances that incorporate this directly into image reconstruction.