5015
Automated MRI k-space Motion Artifact Detection in Segmented Multi-Slice Sequences1Athinoula A. Martinos Center for Biomedical Imaging, Department of Radiology, Massachusetts General Hospital, Boston, MA, United States, 2Department of Radiology, Harvard Medical School, Boston, MA, United States, 3Computer Science and Artificial Intelligence Laboratory, Massachusetts Institute of Technology, Cambridge, MA, United States, 4Harvard-MIT Health Sciences and Technology, Massachusetts Institute of Technology, Cambridge, MA, United States, 5MGH & BWH Center for Clinical Data Science, Mass General Brigham, Boston, MA, United States, 6GE Healthcare, Chicago, IL, United States
Synopsis
Motion artifacts negatively impact diagnosis and the radiology workflow, especially in cases where a patient recall is required. Detecting motion artifacts while the patient is still in the scanner could potentially improve workflow and reduce costs by enabling efficient corrective action. We introduce an algorithm that detects motion artifacts directly from raw k-space in a supervised learning manner in clinically important 2D FSE multi-slice scans, using cross-correlation between adjacent phase-encoding lines as features. This study employs a training approach that uses a motion simulator to generate k-space data with varying levels of motion artifact.
Authorship Information
† and * denote equal contribution.Introduction
Diagnosis from magnetic resonance (MR) images is often difficult due to image artifacts despite continuing improvements in MR hardware. Motion corruption is one of the most common artifacts mentioned in radiology reports and affects about 15% of brain MR exams [1, 2]. Motion artifacts negatively impact the radiology workflow, especially in cases where a patient recall is required. Detecting motion artifacts while the patient is still in the scanner could potentially improve workflow, for example by alerting technicians to artifacts after, or even during a scan acquisition, so that efficient corrective action can be taken. We introduce an algorithm that detects motion artifacts directly from raw k-space in a supervised learning manner in clinically important 2D Fast Spin Echo (FSE) multi-slice scans [3], using cross-correlation between adjacent phase-encoding lines [4] as features. To avoid relying on imbalanced data (i.e., fewer data with severe motion artifact), we investigate a training approach that uses a motion simulator to generate k-space data with varying levels of motion artifact.Methods
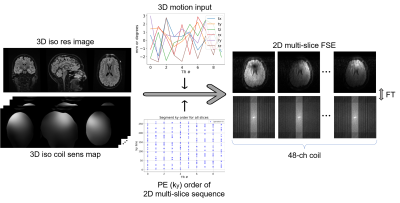
1) Data collection: We collected DICOM and raw k-space data from clinical (outpatient) brain MRI scans at our hospital and de-identified the metadata under an approved Institutional Review Board protocol. We developed a remote data retrieval system that allows for the acquisition of k-space and DICOM paired data without significantly affecting clinical workflow.2) Data: In brief, we acquired 3D isotropic high-resolution data, applied simulated motion, and subsampled the k-space to generate 2D FSE multi-slice data. 3D isotropic resolution Cube T2 FLAIR data (30 studies) were acquired using a 3T GE SIGNA Premier system and 48-channel head coil with the following imaging parameters: TR=6300ms, TE=110ms, FOV=256x230mm2, acquisition matrix=[272, 246], slice thickness=1.0-1.4mm. The motion simulation pipeline produced 2D FSE multi-slice axial T2 FLAIR sequence (ARC acceleration factor=3, TR=10000ms, TE=118ms, FOV=260x260mm2, acquisition matrix=[416, 300], slice thickness=5mm, slice spacing=1mm) from the 3D isotropic data.
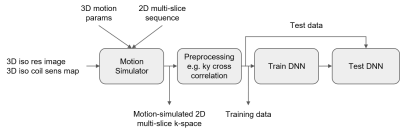
3) Motion Simulation to generate training data: Using knowledge of the sequences’ k-space sampling, a forward model to simulate motion artifacts in 2D multi-slice data has been developed (Figure 2). The simulation takes as inputs: (a) a 3D isotropic image; (b) coil sensitivity maps, estimated with ESPIRiT [5]; (c) head position for the sampling of each k-space segment. The output is multi-channel k-space data where each k-space segment has been sampled from a slice at the supplied head position. This forward model enables simulation of both in-plane and through-plane motion. Rigid-body head motion was used to simulate different levels of motion artifact (no/mild/moderate/severe motion) by controlling the motion parameters. 33600 k-space datasets (30 studies x 28 slices x 4 motion severity x 10 augmentations) were generated, each corresponding to an anatomical slice. The dataset was split into three at the study level. 60% of the studies were used for training, 20% for validation, and 20% for testing.
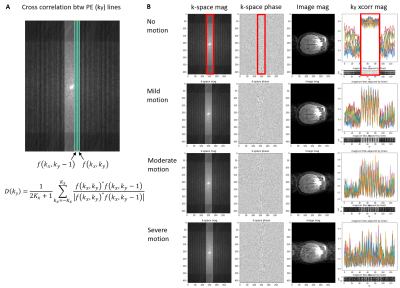
4) Preprocessing: Preprocessing was used to extract motion-related features and reduce data dimension. The underlying idea is to detect inconsistencies in k-space caused by motion. With the hypothesis that data in the neighboring ky phase encoding (PE) lines are not very different unless motion occurs [4], we calculated the normalized cross-correlation between adjacent ky lines (called “ky cross-correlation” in this paper)
$$D(k_y)=\frac{1}{2K_x+1}\sum_{k_x=-K_x}^{K_x}\frac{f(k_x,k_y)^*f(k_x,k_y-1)}{\mid f(k_x,k_y)^*f(k_x,k_y-1) \mid},$$
where $$$f(k_x, k_y)$$$ is 2D k-space and $*$ is the complex conjugate (Figure 3A). This initial investigation used the magnitude of cross-correlation, the center of k-space where it is fully sampled (self-calibrated region), and 12 coil channels out of 48. This process reduces data dimensions from 4 to 2 for each sample. Examples are shown in Figure 3B.
5) Training deep neural networks (DNN): A ResNet-18-like convolutional neural network was used to classify k-space data into 4 classes -- i.e., no/mild/moderate/severe motion. The ResNet-18 was modified to have a single channel and the fully connected layer was modified to output 4 values. Parameters such as kernel size and padding were modified to account for the smaller input shape. Cross entropy loss and Adam optimizer were used. The learning rate started at 0.0005 and decayed exponentially.
6) Testing: The trained network was tested on the test set and the classification performance was measured.
Results
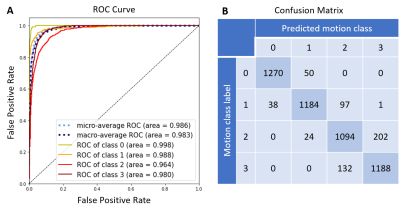
The Cohen's kappa with linear weights was 0.918 and 0.959 when quadratically weighted. The overall accuracy was 0.897. The area under the micro-average ROC curve (AUROC) across 4 classes was 0.986. The model found it easiest to detect data with no motion artifact (AUROC=0.998) and most difficult to detect data with moderate motion artifact (AUROC=0.964; Figure 4).Discussion
The proposed model shows competitive performance to detect motion artifacts in k-space of a segmented multi-slice sequence. We observe that, in the preprocessing (ky cross-correlation), data with greater motion show signal discontinuity (spikes) across PE lines (Figure 3B). Future studies will validate the current model on 2D FSE multi-slice data with real motion, first by doing volunteer study using guided motion and second by collecting patients’ data in our hospital. Further, we plan to test the pipeline on other contrast such as T2 and FLAIR scans.Conclusion
The proposed methods for motion artifact detection in MRI can help improve image quality, reduce patient recall, and enhance radiology workflow.Acknowledgements
We are grateful for support for this research which was provided by GE Healthcare and the National Institutes of Health (U01MH117023, R01 MH123195, R01EB023281, R01EB006758, R21EB018907, R01EB019956, P41EB030006, R56AG064027, R01AG008122, R01AG016495, 1RF1MH123195, R01MH123195, R01MH121885, R01NS052585, R21NS072652, R01NS070963, R01NS083534, U01NS086625, U24NS10059103, R01NS105820, 1S10RR023401, 1S10RR019307, 1S10RR023043, 5U01-MH093765).
References
- Andre JB, Bresnahan BW, Mossa-Basha M, et al. Toward quantifying the prevalence, severity, and cost associated with patient motion during clinical MR examinations. Journal of the American College of Radiology. 2015;12(7):689-95.
- Gedamu EL, Gedamu A. Subject movement during multislice interleaved MR acquisitions: prevalence and potential effect on MRI‐derived brain pathology measurements and multicenter clinical trials of therapeutics for multiple sclerosis. Journal of magnetic resonance imaging. 2012;36(2):332-43.
- Hennig J, Nauerth A, Friedburg HR. RARE imaging: a fast imaging method for clinical MR. Magnetic resonance in medicine. 1986;3(6):823-33.
- Mendes J & Parker DL. Intrinsic detection of motion in segmented sequences. Magnetic resonance in medicine. 2011;65(4):1084-9.5.
- Uecker M, Lai P, Murphy MJ, et al. ESPIRiT—an eigenvalue approach to autocalibrating parallel MRI: where SENSE meets GRAPPA. Magnetic resonance in medicine. 2014;71(3):990-1001.
Figures